|
|
|
| P R I N T E R - F R I E N D L Y F O R M A T | Return to Article |
D-Lib Magazine
November/December 2014
Volume 20, Number 11/12
Towards a Marketplace for the Scientific Community: Accessing Knowledge from the Computer Science Domain
Mark Kröll, Stefan Klampfl and Roman Kern
Know-Center GmbH, Graz, Austria
{mkroell, sklampfl, rkern}@know-center.at
doi:10.1045/november14-kroell
Abstract
As scientific output is constantly growing, it is getting more and more important to keep track not only for researchers but also for other scientific stakeholders such as funding agencies or research companies. Each stakeholder values different types of information. A funding agency, for instance, might be rather interested in the number of publications funded by their grants. However, information extraction approaches tend to be rather researcher-centric indicated, for example, by the type of named entities to be recognized. In this paper we account for additional perspectives by proposing an ontological description of one scientific domain — the computer science domain. We accordingly annotated a set of 22 computer science papers by hand and make this data set publicly available. In addition, we started to apply methods to automatically extract instances and report preliminary results. Taking various stakeholders' interests into account as well as automating the mining process represent prerequisites for our vision of a "Marketplace for the Scientific Community" where stakeholders can exchange not only information but also search concepts or annotated data.
Keywords: Scientific Stakeholders, Marketplace, Computer Science Domain, Scientific Publications, Knowledge Acquisition
1. Introduction
The scientific community produces output by publishing their activities and achievements, for instance, in journal articles. As this scientific output is constantly growing, it is getting more and more difficult to keep track not only for researchers but also for other scientific stakeholders such as funding agencies, research companies or science journalists. Each stakeholder values different types of information. So far, research assistants such as BioRAT [2] or FACTA [9] are often researcher-centric, i.e. scanning scientific publications for interesting facts in their respective domain. In contrast, a funding agency might be interested in grant information in combination with the research team, while a research company might be more interested in which algorithm works best for a specific data set.
We account for these different perspectives by ontologically describing one scientific domain — the computer science domain. Guided by questions such as "What is considered valuable information from a stakeholder's viewpoint", we introduce (i) stakeholder specific categories such as "Funding Information" or "License" and (ii) more general categories such as "Research Team" or "Performance". We then add linkage information between these categories to describe factual knowledge in form of triples. Linking these facts together into information blocks creates value, for example, by briefly summarizing scientific content. Information blocks can vary from stakeholder to stakeholder, for instance, a research company might be interested in following information block: {"Task", "Algorithm", "Software", "License"} while a graduate student might be interested in the following one: {"Scientific Event", "Task", "Corpus", "Algorithm", "Performance", "Numeric Value"}. We consider these categories and their combinations as useful and as a prerequisite for providing an interactive marketplace for researchers where scientific stakeholders can exchange not only information but also manually annotated data of high quality.
According to our ontological description we manually annotated a data set containing 22 computer science publications. Annotating these scientific publications served two purposes. The first one was to refine and update our ontological description by examining real-world data and thus to get a better understanding of instances to expect and how to extract them. The second one was to create a small data set for testing and evaluating learnt models to automate instance as well as relation extraction. In a first step we applied simple extraction methods such as regular expressions and gazetteer-based approaches for five selected categories. Finally, we report our observations with respect to category characteristics and discuss achieved results as well as future steps.
2. Ontological Description of the Computer Science Domain
Scientific publications constitute an extremely valuable body of knowledge whose explication aids scientific processes including state-of-the-art research, research comparison or re-usage. Explicating scientific knowledge includes extracting facts from large amounts of data [7].
However, efforts to automatically extract knowledge from scientific domains so far have rather been researcher-centric. Medical entity recognition [1] focuses on classes such as "Disease", "Symptom" or "Drug". In bioinformatics [10], the focus is on identifying biological entities, for example, instances of "Protein", "DNA" or "Cell Line", and extracting the relations between these entities as facts or events. Departing from a mere content-level, Liakata et al. [4] introduced a different approach by focusing on the discourse structure to characterize the knowledge conveyed within the text. For this purpose, the authors identified 11 core scientific concepts including "Motivation", "Result" or "Conclusion". Ravenscroft et al. [6] present the Partridge system which automatically categorizes articles according to their types such as "Review" or "Case Study". Teufel et al. [8] use rhetorical elements of a scientific article such as "Aim" or "Contrast" to generate summaries.
In a similar attempt, we ontologically describe the computer science domain with respect to various stakeholders by taking their motivations into account to scan scientific publications. Scientific stakeholders we considered during the designing process include:
- Researchers who are interested in (i) keeping up-to-date with activities and achievements in their respective and/or related field; (ii) conducting literature surveys for writing a related work section or getting ideas for research questions; (iii) getting quickly up-to-speed when acting as a reviewer.
- Companies who aim (i) to identify applicable techniques for a particular task, for example, which algorithm or software can be used (license constraints) (ii) to get automatically informed about newly introduced methods in certain fields or (iii) to gain knowledge in a particular field.
- Funding agencies who are interested in the number of publications which are funded by their grants.
- Science journalists who want to quickly get an overview about the state-of-the-art in a certain field.
To account for these perspectives we introduce 15 categories which are described in Table 1.
Table 1: Categories of the Computer Science Domain.
| Category | Description |
| Domain | includes fields or subfields such as "Machine Learning" or "Information Extraction". |
| Corpus | encompasses data sets such as "British National Corpus", "GENIA" or "MNIST". |
| Scientific Event | includes conferences or challenges such as "2004 Bio Creative" or "SemEval". |
| Algorithm | encompasses algorithms or methods such as "PCA" or "Support Vector Machine". |
| Performance | contains standard metrics such as "F-Measure" or "accuracy". |
| Website | such as "mybarackobama.com", "MyBo" or "Twitter". |
| Numeric Value | contains not only numeric expressions (75%) but also expressions such as "2 orders of magnitude" or range information. |
| Parameter | such as the SVM penalty parameter "C". |
| Task | is meant to capture a general task of the community or the paper's task itself such as "POS tagging" or "noun-phrase chunking". |
| Software | encompasses software, systems, tools, programming languages ("C++") or libraries ("Weka" or "Stanford Parser"). |
| Artifact | exists in the real world such as "Pentium 4 processor". |
| Format | describes encodings such as "UTF-8". |
| License | describes the directives of using a software system as is defined in licenses such as "GPL". |
| Research Team | is not restricted to a team or a group, yet omits references. It can also encompass an entire lab, institute or university such as "Turing Center" or "Stanford NLP Laboratory". |
| Funding Information | provides grant information such as "NSF Fellowship" or "NSF grant #IIS-0711348". |
In Figure 1 we present an ontological description of the computer science domain to explicate knowledge and thereby making it accessible and usable. The domain model contains categories as well as linkage information. Relation categories include "Achieve", "AppliedTo", "Develop", "Generate", "Get", "Have, "RelevantFor", "Require", "TrainedOn", "Use" and "WorkIn". Relations between categories are required to describe factual knowledge that can also aid the process of generating summaries of scientific content [5].
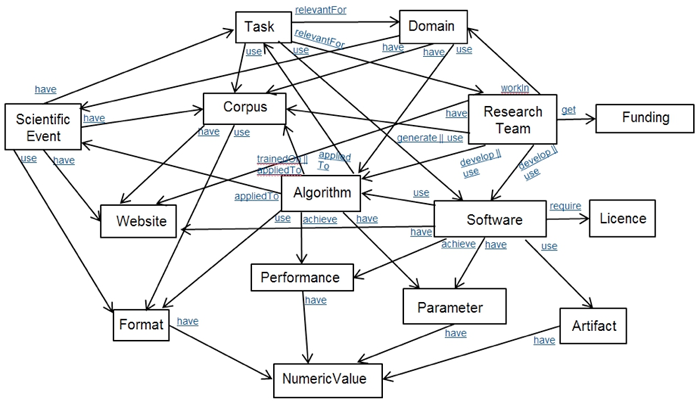
Figure 1: Ontological Description of the Computer Science Domain.
Figure 2 illustrates an annotation example with reference to our ontological description. The annotation example shows a potential population of the information block: {"Scientific Event", "Task", "Corpus", "Algorithm", "Performance", "Numeric Value"} in the introduction. This kind of factual knowledge might be interesting, for instance, for graduate students compiling state-of-the-art. To generate real value, the extraction process needs to be automated to handle larger amounts of scientific publications — the proposed domain model in combination with the manually annotated data set represents a first step into this direction.
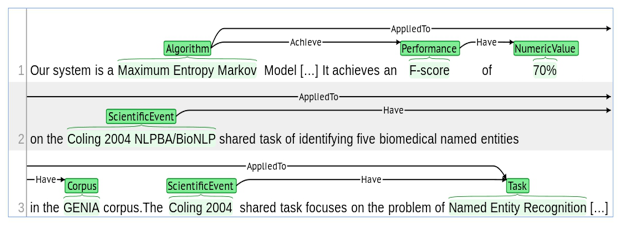
Figure 2: Annotation Example using the proposed Ontological Description of the Computer Science Domain. We used the BRAT annotation tool in this example.
Finally, we point out additional benefits of an ontological description including (i) support for reasoning systems by using constraints or (ii) guidance for automated acquisition processes. In addition, we refrain from arguing completeness of the ontological description, yet, we firmly believe that the proposed categories plus their relationships capture knowledge which is beneficial for various scientific stakeholders.
3. Data Set
To automatically extract the textual content from scientific publications, we developed a processing pipeline [3] that applies unsupervised machine learning techniques in combination with heuristics to detect the logical structure of a PDF document. For annotation purposes we used the BRAT rapid annotation tool which provided us with an easy-to-use annotation interface. The annotation process was preceded by a setup phase where we examined a couple of papers to create a common understanding of the categories. After that, two of the authors manually annotated 22 computer science papers (11 papers each) from various subdomains including information extraction, computer graphics or hardware architecture to broaden the instance variety. In total the data set contains 5353 annotations — 4773 category annotations and 580 relation annotations. Table 2 provides statistics about the unique instances per category.
Table 2: Number of (unique) instances per category.
| Category | #instances (unique) | Category | #instances (unique) | |
| Domain | 86 (35) | Numeric Value | 991 (593) | |
| Corpus | 187 (51) | Parameter | 102 (39) | |
| Scientific Event | 35 (21) | Task | 522 (200) | |
| Algorithm | 734 (200) | Software | 944 (192) | |
| Performance | 800 (187) | Artifact | 93 (38) | |
| Website | 87 (29) | Format | 125 (32) | |
| Funding Information | 37 (30) | License | 4 (3) | |
| Research Team | 26 (21) |
The zip file containing the data set can be downloaded here. Beside the .txt and respective .ann files the zip container includes an annotation.conf file for the BRAT viewer as well as information about the annotated publications.
4. Automated Approach
The categories introduced in Table 1 exhibit different characteristics which can be taken into account to select automated (learning) methods. Some of these categories such as "Corpus", "Performance" or "Format" resemble named entity categories in the classical sense such as "Person" or "Location". Instance additions are rather the exception than the rule making them nearly closed. This closeness property makes them accessible to simple gazetteer-based approaches. Categories such as "Numeric Value" or "Funding Information" encompass characteristical instances, for instance, containing digits, occurring at certain positions or in certain sections of the publication. Such characteristics can be exploited by regular expressions. Categories such as "Algorithm" or "Task" contain a broad variety of instances. For example, instances of the "Task" category include noun phrases such as "document analysis" as well as verb phrases such as "acquiring common sense knowledge" making an automated recognition challenging.
In the following section we apply regular expressions and gazetteer-based approaches to our dataset to automatically extract instances from following categories: "Algorithm", "Performance", "Numeric Value", "Corpus" and "Funding Information".
4.1 Preliminary Results
For the categories "Algorithm", "Corpus" and "Performance" we applied a gazetteer-based approach and crawled respective lists on-line. Wikipedia, for instance, offers some lists containing algorithms used in the computer science domain. We manually reviewed acquired category instances and discarded (i) incorrect entries, (ii) too general entries, for instance, "scoring algorithm" and (iii) ambiguous entries such as "birch" which can relate to either the clustering algorithm or the tree. Our gazetteer lists contain following number of entries: 1193 for the category "Algorithm", 105 for the category "Corpus" and 85 for the category "Performance". To pre-process the textual content, we applied (i) tokenization, (ii) sentence splitting, (iii) normalization & stemming and (iv) part-of-speech tagging. The gazetteer-based annotator focuses on stems and matches the longest sequence only so that a gazetteer entry "support vector machine" would match the plural "support vector machines" but not the phrase "support vector".
After scanning through instance characteristics of the categories "Numeric Value" and "Funding Information", we opted for a regular-expression based approach. Regular expressions to extract "Numeric Value" instances were designed to include instances such as such as "60 million", "50 MHz" and "2.5 orders of magnitude" and neglecting numbers which are part of references or instances such as "Figure 1". Funding information such as grants is almost always provided at the end of a paper exhibiting a clear structure, i.e. typical a noun phrase containing digits, indicated by trigger phrases such as "<supported | funded | financed><by>". Recognition results for these five selected NE categories are shown in Table 3.
Table 3: Precision, Recall and F-Scores for selected categories.
| Category | Precision | Recall | F1 - Score |
| Algorithm | 0.68 | 0.34 | 0.45 |
| Corpus | 0.94 | 0.50 | 0.65 |
| Performance | 0.80 | 0.49 | 0.61 |
| Numeric Value | 0.76 | 0.87 | 0.81 |
| Funding Information | 0.79 | 0.70 | 0.74 |
To calculate precision, recall and subsequently the F1-score, we counted correctly, partially correct and incorrectly matched annotations as well as the number of missing (not in the evaluation set) and spurious annotations (not in the ground truth). Precision is calculated as follows: (correct + ½ partially correct) / (correct + partially correct + incorrect + spurious). Recall is calculated as follows: (correct + ½ partially correct) / (correct + partially correct + missing).
4.2 Discussion
The result values in Table 3 corroborate our intuition that the introduced categories exhibit different characteristics and therefore benefit from different extraction approaches. While rather closed or well-defined categories such as "Performance" and "Funding Information" react well to simpler approaches, other more open categories such as "Algorithm" show the need for more sophisticated approaches. The results can serve as a baseline for learning approaches such as maximum entropy models or conditional random fields which are widely used in the field of named entity recognition. However, 22 annotated papers do not provide enough data for training these models. We thus intend to compile larger training sets (i) using manually annotated instances to enlarge our gazetteer lists as well as (ii) using automatically annotated instances as candidates for training the models.
5. Conclusion
In this paper we create awareness of different stakeholder perspectives when processing and accessing scientific data. Taking stakeholders' interests into account represents a prerequisite for an interactive marketplace which serves the scientific community to access an extremely valuable body of knowledge. For that purpose we ontologically describe one scientific domain, the computer science domain and accordingly annotated 22 publications. In a first step we apply simple methods such as regular expressions and gazetteer-based approaches to automatically extract instances of five selected categories. Future steps include (i) compiling larger sets of training data, (ii) applying extraction algorithms which take, for instance, sequence information into account as well as (iii) detecting relations between the categories.
Last but not least this paper contributes to the vision of establishing a commercially oriented ecosystem, i.e. a marketplace for scientific stakeholders to interact. This interaction, in our opinion, generates not only high-quality knowledge, but also commercial value for all participants.
Acknowledgements
We thank the reviewers for their constructive comments. The presented work was developed within the CODE project funded by the EU FP7 (grant no. 296150). The Know-Center is funded within the Austrian COMET Program — Competence Centers for Excellent Technologies — under the auspices of the Austrian Federal Ministry of Transport, Innovation and Technology, the Austrian Federal Ministry of Economy, Family and Youth and by the State of Styria. COMET is managed by the Austrian Research Promotion Agency FFG.
References
[1] Abacha, A. and Zweigenbaum, P. 2011. Medical entity recognition: a comparison of semantic and statistical methods. BioNLP 2011 Workshop. Association for Computational Linguistics.
[2] Corney, D., Buxton, B., Langdon, W. and Jones, D. 2004. BioRAT: extracting biological information from full-length papers. Bioinformatics, 20. http://doi.org/10.1093/bioinformatics/bth386
[3] Klampfl, S. and Kern, R. 2013. An Unsupervised Machine Learning Approach to Body Text and Table of Contents Extraction from Digital Scientific Articles. Research and Advanced Technology for Digital Libraries.
[4] Liakata, M., Saha, S., Dobnik, S., Batchelor, C. and Rebholz-Schuhmann, D. 2012. Automatic recognition of conceptualization zones in scientific articles and two life science applications. Bioinformatics 28 (7).
[5] Liakata, M., Dobnik, S., Saha, S., Batchelor, C. and Rebholz-Schuhmann, D. 2013. A discourse-driven content model for summarising scientific articles evaluated in a complex question answering task. Conference on Empirical Methods in Natural Language Processing.
[6] Ravenscroft, J., Liakata, M. and Clare, A. 2013. Partridge: An Effective System for the Automatic Classification of the Types of Academic Papers. AI-2013: The Thirty-third SGAI International Conference. http://doi.org/10.1007/978-3-319-02621-3_26
[7] Seifert, C., Granitzer, M., Höfler, P., Mutlu, B., Sabol, V., Schlegel, K., Bayerl, S., Stegmaier, F., Zwicklbauer, S. and Kern, R. 2013. Crowdsourcing fact extraction from scientific literature. Workshop on Human-Computer Interaction and Knowledge Discovery.
[8] Teufel, S. and Moens, M. 2002. Summarizing scientific articles: experiments with relevance and rhetorical status. Computational Linguistics 28 (4).
[9] Tsuruoka, Y., Tsujii, J. and Ananiadou, S. 2008. FACTA: a text search engine for finding associated biomedical concepts. Bioinformatics 24(21). http://doi.org/10.1093/bioinformatics/btn469
[10] Zweigenbaum, P., Demner-Fushman, D., Yu, H., and Cohen, K. 2007. Frontiers of biomedical text mining: current progress. Briefings in Bioinformatics, 8(5). http://doi.org/10.1093/bib/bbm045
About the Authors
 |
Mark Kröll is a senior researcher at the Know-Center with expertise in the fields of knowledge acquisition, knowledge management and natural language processing. He received both his Master's degree in Telematics as well as his PhD degree in Computer Science at Graz University of Technology. |
 |
Stefan Klampfl is a postdoctoral researcher at the Know-Center, with expertise in the fields of machine learning and data analysis. He received both his DI degree (in Telematics; equivalent to an MSc in information and communication technology) and his PhD (in computational neuroscience) at Graz University of Technology. In his PhD thesis he investigated unsupervised machine learning algorithms and their role in information processing in the brain, and analyzed biological data using methods from machine learning and information theory. While writing his thesis, he contributed to a number of high-impact conference and journal publications. |
 |
Roman Kern is the division manager of the Knowledge Discovery area at the Know-Center and senior researcher at the Graz University of Technology, where he works on information retrieval and natural language processing. In addition he has a strong background in machine learning. Previous to working in research he gained experience in industry projects at Hyperwave and Daimler. There he worked as project manager, software architect and software engineer for a number of years. He obtained his Master's degree (DI) in Telematics and his PhD in Informatics at the Technical University of Graz. After his studies he worked at a start-up company in the UK as a Marie Curie research fellow. He participates in a number of EU research projects, where he serves as coordinator and work package leader. He manages a number of large research and development projects in cooperation with the industry. He also gives lectures at the Technical University of Graz for Software Architecture, Knowledge Discovery and Data Science. He also serves as supervisor for Bachelor, Master and PhD students. He published over 40 peer-reviewed publications and achieved top rank results in international scientific challenges like CLEF, ACL SemEval. Most of his publication work can be accessed online here. |
|
|
|
| P R I N T E R - F R I E N D L Y F O R M A T | Return to Article |