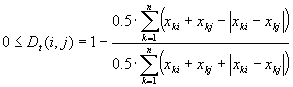
1Federal Research Centre for Forestry and Forest Products,
Institute for Forest Genetics and Forest Tree Breeding,
2Centre for Applied Plant Molecular Biology, AMP I,
Institute for General Botany,
3INRA, Station de Recherches Forestières Guayane,
*Corresponding author: Email: ziegenha@aixh0001.holz.uni-hamburg.de
Abstract
Within populations of forest tree species, highly polymorphic genetic markers are supposed to enable the reconstruction of family relationships. This pilot study is on validating AFLPs as DNA markers for differentiation among full- and half-sib-family relationships in the broad-leaved tree species oak (Quercus spp.). For this purpose, the DIG-labelled AFLP technique was adapted to oaks and Mendelian inheritance analysis conducted by segregation analysis of polymorphic bands among parents and their full-sib progeny using three different PECs. The polymorphic DNA fragments were mainly found to follow a dominant mode of gene action. Pairwise Tanimoto distances were computed both within a subset of full-sibs as well as within a subset of half-sibs and later on compared. First results reveal different distributions of Tanimoto distances between the two subsets. However, strong overlapping of frequencies occurred in one of nine distance classes. The potential of dominant AFLP markers for reconstruction of family structures is discussed with regard to spatial genetic analysis.
Key-words: trees, Quercus spp., DIG-AFLPs, chemiluminescence, inheritance analysis, genetic distance, family relationships
Abbreviations: AFLP (Amplified Fragment Length Polymorphism),
PEC
(Primer-Enzyme-Combination), DIG (DIG-Oxigenine-based chemiluminescent
detection system according to Boehringer Mannheim, Germany),
AFLP®
(AFLP-Trademark,
Keygene, Wageningen, The Netherlands).
Introduction
In the forest tree genus oak, highly polymorphic nuclear microsatellites have been demonstrated to be powerful codominant DNA markers for parentage analysis and thus for reconstruction of the mating system and/or gene flow (Dow and Ashley 1996, Lexer et al. 1997, Streiff et al. 1999). Furthermore, in oak species, microsatellite markers were successfully applied for verifying/falsifying maternal half-sib relationships from subsets of just a few seeds (Lexer et al. 1999).
What are the perspectives for end-users, e.g. in practical forest management? By assessment of the degree of kinship, e.g. family structures, the effects of forest management on the mating system of a tree species will be better understood. Using the data for simulation, the effects may then be estimated over a few generations (Degen et al. 1996). Another application is examined in the above mentioned study by Lexer et al. (1999, this Compendium): This category of highly polymorphic DNA markers is shown to be a new and promising tool for controlling family relationships within charges of forest reproductive material harvested for trade.
Nuclear microsatellites have one disadvantage: Their development is time and thus money consuming in any species or genus. In contrast, AFLP technology is a universal marker technology. It is therefore worthwhile to test this fingerprint technology for its capacity in comparison with nuclear microsatellites. A comprehensive evaluation of these two DNA marker categories is given in the review article by Robinson and Harris (1999, this Compendium). These authors mainly consider the use of microsatellites and AFLPs in plant population genetics and phylogenetic analyses. For oaks, an experimental approach for testing the applicability of AFLPs in a comparative parentage analysis is presented by Gerber et al. (1999, this Compendium).
Here, a pilot study is presented on validating AFLPs as DNA markers
for differentiating between full- and half-sib family relationships in
the broad-leaved tree 'oak' (Quercus spp.). For this purpose, as
one of the goals of the project, non-radioactively labelled AFLP technology
was adapted to oaks in order to provide a protocol for laboratories that
have neither isotopes nor automated sequencing equipment. In a next step,
a subset of full-sibs as well as a subset of half-sibs were determined
from a single tree progeny by means of microsatellite markers. The full-sib
family was the basis for a comprehensive inheritance analysis checking
three different PECs. In a last step, AFLP banding patterns from full-
and half-sibs were analysed for pairwise Tanimoto distances. The question
that will be later on discussed is: Do AFLP markers provide a reliable
tool for estimating the degree of kinship within a tree population?
Material and Methods
Determination of an oak half-sib and an oak full-sib family from a single tree progeny
Oak seedlings were sampled that had been grown from acorns collected beneath one adult pedunculate oak tree (Quercus robur L.) (the putative seed parent) in the Arboretum of the Institute for Forest Genetics and Forest Tree Breeding in Grosshansdorf. Using nuclear microsatellites as markers for paternity analysis, it was possible to reconstruct subsets of half- and full-sib seedlings from the single tree progeny. For this purpose 45 adult oak trees of the Arboretum, including the seed parent, and a total of 116 seedlings were analysed at three to four microsatellite loci. Based on these data, in a first step those seedlings were excluded from subsequent analyses that did not share any allele at each of three microsatellite loci with the seed parent. The remaining data were subjected to paternity analysis. On the basis of three microsatellite loci, a putative pollen parent could be assigned to the seedlings with an average exclusion percentage of 91. Excluding selfs, the seedlings were tentatively divided into full-sib families, each with a different pollen parent. The maximum size of these tentative full-sib families turned out to be 35. These 35 candidate full-sibs were then analysed at a fourth microsatellite locus. The results finally revealed that 33 of them were unambiguous full-sib seedlings of the seed parent and their respective pollen parent. Furthermore, the data set were screened for the maximum number of seedlings representing a 'true' half-sib-family, where each member has a different pollen parent. 11 seedlings were the maximum number that could be determined to be half-sibs of the same seed parent at the above-mentioned level of exclusion (91%). See below for the methods in detail.
DNA extraction:
50 - 100 mg (fresh weight) leaf material was homogenised in Eppendorf tubes using a shaking mill (Retsch GmbH, Haan, Germany; Ziegenhagen et al. 1993) and total DNA extracted according to Dumolin et al. (1995) with slight modifications including a final treatment with 0.5 µg RNaseA (Boehringer Mannheim, Germany) at 37° C for 30 minutes.
Microsatellite analysis:
The following microsatellite loci as characterised by Steinkellner et al. (1997) were analysed: ssrQpZAG1/5, ssrQpZAG36, ssrQpZAG9, ssrQpZAG104.
PCR amplification: PCR cocktail and cycle profile were designed according to Streiff et al. (1998). PCR was run in the Touch-DownTM Thermal System (Hybaid Limited, Teddington, UK). Separation and staining of PCR products: The PCR products were pre-treated according to Streiff et al. (1998) and run in a 6% denaturating polyacrylamide gel (Rotiphor 40, 38:2 alcrylamide:bisacrylamide, Roth, Karlsruhe, Germany), using a sequencing gel apparatus (S2 Gibco BRL, Life Technologies, Eggenstein, Germany). The gels were run in 1xTBE buffer adjusted to pH 8.3 at 2000 V for 2.5 to 3 h. Silver staining of the gels was performed according to Streiff et al. (1998).
Paternity analysis:
Paternity analysis was performed by the Computer-programme FAMILIA,
v. 1.0 (Degen 1996, unpublished). This programme operates by paternity
exclusion and on the assumption of a spatially restricted gene flow.
Performance of DIG-AFLP technology and analysis of banding patterns
DIG-Oxigenine protocol
Total DNA from the parents and their 33 full-sib offsprings as well as the total DNA of the seed parent and its 11 half-sib offsprings was analysed for AFLP banding patterns. DIG-labelled AFLP technology was adapted to oaks using a combination of different protocols. The basic protocol is that by the patent holder for AFLP® (Vos et al. 1995). Our DIG-Oxigenine approach is a modification of a protocol by Gebhardt (1995, unpublished, MPI, Köln, Germany) that was optimised for maize by Brettschneider (1996). Details of our protocol will be published elsewhere (Ziegenhagen et al., submitted).
Three different PECs were chosen that are assumed to follow different profiles of sampling within the genome (more random-like or more cluster-like profiles, Peleman, personal communication):
Eco RI + AAC - Mse I + ACT
Hind III + ACC - Mse I + ACT
Pst I + CCA - Mse I + ATA
Inheritance analysis:
Inheritance analysis was conducted on the full-sib family taking the DNA banding patterns, each as generated by the three different PECs. The banding patterns were screened for polymorphic bands, these band positions assigned with discrete values from '1' to 'n' ranging from the smallest to the greatest length of the DNA fragments. In this way, the gel lanes of the individual AFLPs of seed and pollen parent followed by the 33 full-sibs were translated into columns of three Excel tables. The latter were imported into the computer programme CoTrix (Degen 1996). By means of this programme the banding patterns were digitised into a 1/0 matrix (present/absent band at the relevant band position) which is the basis for segregation analysis. From the obtained segregation data, the programme reconstructs the most probable parental combination according to the possible six different cases of combinations. Test of null hypothesis is done by means of a G-Test which is included in the programme (for the six cases, see Degen 1996 and below).
Distance analysis:
Pairwise estimation of genetic distances between individuals were done by computing similarity/dissimilarity of AFLP banding patterns. The Tanimoto distance measure was used for pairwise comparison of the individual banding patterns within both subsets, the full-sibs and the half-sibs. This comparison of banding patterns was done on the Pst/Mse PEC only.
The Tanimoto distance was calculated by the programme CoTrix as follows (formula in: Deichsel and Trampisch 1985):
where
k = band position,
n = number of different band positions,
xki = value of the 1/0 matrix of band position k of the individual i, and
xkj = value of the 1/0 matrix of band position k of the individual
j.
Results
Adaptation of DIG-labelled AFLP technology
DIG-labelled AFLP technology was successfully adapted to oak and provided distinct and reproducible banding patterns. The usage of all three PECs resulted in highly polymorphic patterns, the PECs Eco/Mse and Hind/Mse exhibiting a greater number of bands and a greater number of polymorphic bands than the PEC Pst/Mse. Figure 1 gives an example of DIG-detected AFLP banding patterns in a subset of the analysed oak full-sib family.

Figure 1: DIG-detected AFLP patterns of a full-sib oak family. Section of a gel demonstrating the patterns by using PEC Pst/Mse. M = seed parent, F = pollen parent (M and F were loaded in replicates), KB = Molecular Size Standard V (Boehringer, Mannheim, Germany).
|
No. of bands scored |
(six cases, I to VI) |
|||||
|
|
|
|
|
|
|
|
|
|
||||||
|
46 |
|
|
|
|
|
|
|
48 |
|
|
|
|
|
|
|
28 |
|
|
|
|
|
|
Table 1: Results of segregation analysis of polymorphic bands among parents and full-sib progeny. As defined in CoTrix (Degen 1996), the parental combinations are as follows:
| I = | parental combination I: both parents are different homozygotes |
| II = | parental combination II: both parents are different heterozygotes |
| III = | parental combination III: both parents are different heterozygotes and share one allele |
| IV = | parental combination IV: both parents have the same heterozygous genotype |
| V = | parental combination V: one parent with heterozygous, the other with homozygous genotype: the homozygote shares one allele of the heterozygote |
| VI = | parental combination VI: one parent with heterozygous. the other with homozygous genotype: the genotypes do not share any allele |
Inheritance analysis
Table 1 gives the results of segregration analysis. For all three PECs, the parental combination V was predominantly reconstructed. Only once was the parental combination IV also reconstructed, however at a low level of significance (ns=1.05). From all combinations in case V, those band combinations were regarded as candidates for codominance that were reconstructed at a level of significance of ns<0.05. For the obtained candidate combinations (for numbers, see Table 1) the hypothesis of 'codominance' cannot be rejected. Thus at first glance, there is an unexpectedly high number of band combinations that significantly fit with codominant mode of gene action. However, among the parental combination V, all band combinations share one band with another significant combination. In sum, only one candidate combination from case IV out of a total of 122 polymorphic bands indeed reflects a low proportion of codominant AFLP marker bands. This confirms the already known predominantly dominant mode of gene action in AFLP banding patterns (see discussion).
Tanimoto distances
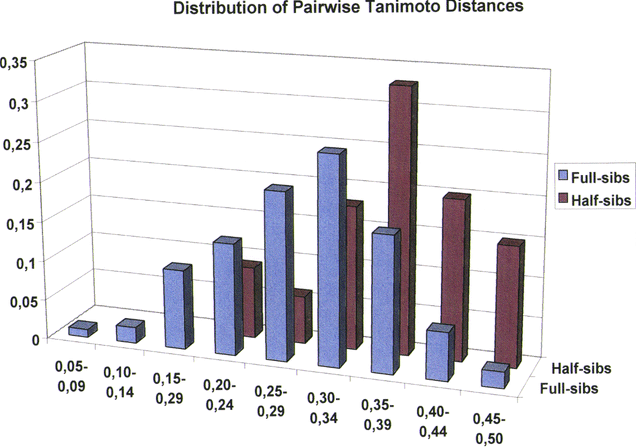
Results of the pairwise Tanimoto distance analysis are summarised in Figure 2. The figure demonstrates the relative occurrences of pairwise Tanimoto distances within 9 different distance classes.
Whereas the parents of the full-sibs are characterised by Tanimoto distance of 0.55 (data not shown), all members of either the full-sib or the half-sib family are less distant to each other. The values of the Tanimoto distance are all < 0.50 except three pairwise combinations of half-sibs that achieved a maximum value of 0.50. As can be judged from Figure 2, the Tanimoto distances follow more or less a Gauss distribution and furthermore it is observed that the distribution is different between the two subsets. For the half-sibs there is a visible shift of the curve towards greater Tanimoto distances. In this subset, as opposed to the full-sibs, the first three distance classes are not represented. The half-sibs only start at a Tanimoto distance of 0.20. In the first three classes, a distinct differentiation between the subsets of full- and half-sibs is possible. On the other hand, there is a strong overlapping of frequencies in distance class No. 6 (0.30-0.34). Here, half- and full-sibs cannot be distinguished, not even by frequencies.
Discussion
In our study we succeeded in adapting non-radioactive AFLP technology to the forest tree 'oak' (Quercus spp.). Our protocol (Ziegenhagen et al., submitted) is in line with other chemiluminescent protocols that have already been published for different organisms including herbaceous plant species (e.g., Lin et al. 1997, Vrieling et al. 1997). Recently, De Greff et al. (1998) reported on a special chemiluminescent AFLP technique applied on sessile oak (Quercus petraea (Matt.) Liebl.). They used the kit 'AFLP Analysis System I' by Gibco BRL (Life Technologies).
Our protocol revealed the reproducibility of banding patterns when applied to independent DNA extractions from the same individual. Also, in a network of European laboratories, AFLPs turned out to be extremely highly reproducible when working on the same DNA with the same detection system (Jones et al. 1998). Working on the same DNA samples, it could also be shown that there is still a remarkable reproducibility between two basically different AFLP detection techniques: the DIG detected AFLP patterns (our laboratory, Grosshansdorf) and the patterns detected by automated sequencing equipment (INRA, Bordeaux) were very close to each other (Kremer, personal communication).
Since AFLPs were introduced as a multi-locus fingerprinting technique in plants (Vos et al. 1995), this technique is being extensively exploited as DNA markers in numerous genetic analyses. Robinson and Harris (this Compendium) give a comprehensive evaluation of this marker as applied in population genetics and phylogenetic studies. Moreover, the potential of AFLPs has been demonstrated for genome mapping, QTL analysis, and for identifying genomic regions involved in disease resistance (e.g., Cervera et al. 1996, Debener and Mattiesch 1999).
As in other studies, also in our analysis this multi-locus approach resulted in highly polymorphic DNA banding patterns. It is of interest to test its capacity for analysing population genetic processes at the individual level or at the within-population level. At this scale and in forest trees, AFLPs may be a desired alternative to nuclear microsatellites in view of the universal applicability of AFLP throughout a large range of forest tree species. The usage of DNA polymorphisms as markers in population genetics necessarily needs knowledge on the mode of inheritance. Thus we determined an oak full-sib family from a single tree progeny using nuclear microsatellite markers. By means of this 'reconstructed controlled cross' we performed inheritance analysis. This is supposed to be difficult in multi-locus DNA fingerprints, as band combinations representing different alleles of the respective locus may not necessarily be neighbouring bands in the gel (Degen et al. 1995). However, the computer programme CoTrix (Degen 1996) enabled a complete segregation analysis of the digitised banding patterns. On the basis of a 0/1 segregation of the bands, only one combination out of 122 bands was detected that followed a codominant mode of gene action. So far, many other combinations in our study that were found to fit with codominance could not be explained, as these combinations always shared one band with another candidate combination. Still, codominance cannot be ruled out in these cases, as DNA fragments of identical size are not necessarily fragments of identical sequence. An answer to this question can only be given by sequence analysis. In sum, our results as obtained by this 'qualitative' (absence/presence of bands) inheritance analysis are in agreement with other published results: With respect to the fragments originating from nuclear DNA (also, to some extent fragments from organelle DNA may be included in the patterns) AFLPs are more or less dominant genetic markers (e.g., Prabhu and Gresshoff 1994).
In spite of their not being codominant markers, we tested the capacity of AFLPs for genetic differentiation among full- and half-sibs of oaks. First results of this pilot study reveal that pairwise Tanimoto distances are differently distributed within these two subsets. A distinct differentiation may be possible in the 'low-frequency' distance classes. In contrast, strong overlapping of frequencies in a 'high-frequency' distance class is supposed to prevent an overall differentiation. This effect of overlapping may be due to the prevailing dominant mode of inheritance, at least as concluded from 0/1 segregation analysis. Also, non-homology or co-migration may be a widespread phenomenon with this marker, and it may introduce a bias into genetic distance analysis (Robinson and Harris, this Compendium). So far, there is only little knowledge on the proportion of codominant marker bands when, instead of presence/absence, the intensity of the bands is used to infer the homo- or heterozygosity-status (Castiglione et al. 1999).
Furthermore, the pilot study needs to be completed. Further analyses should include more PECs that are known to sample different genomic regions and should be conducted on an increased sample size for both the two different subsets and members of the subsets. By this, our first results may be strengthened and a definite answer be given to the initial question.
Since according to our results, a distinct differentiation between full- and half-sib families seems difficult, as is consequently an assessment of the degree of kinship within randomly sampled subsets of a population, what about other possible purposes for this DNA marker in population genetics?
First, there is good evidence for the usefulness of this marker for
paternity analysis in case nuclear microsatellite markers are not available.
Recently, Gerber et al. (this Compendium)
demonstrated the capacity of AFLPs for parentage analysis in oaks with
exclusion percentages almost as high as those achieved by nuclear microsatellites.
Furthermore by AFLPs, De Greef et al. (1998) explored numerous single
plus tree progenies of oak for the level of within-family genetic variation.
They found it to be higher when assessed with AFLPs than with other DNA
markers and assumed this level of variation to be an appropriate basis
for breeding purposes. Degen
et al. (1999) determined a strong spatial
genetic differentiation of a tropical tree species using AFLPs and interpreted
it in terms of family structures. Ecological genetic studies revealed a
correlation between AFLP variation and the variation of the considered
ecophysiological trait (e.g., Pakniyat et al. 1997). The
usage of AFLPs in the latter two studies addresses an old request for markers
that may differentiate between stochastic population genetic processes,
such as the mating system, and processes due to selection. Hence, in spatial
genetic analyses it may be of future interest to sort out from the patterns
polymorphic bands that mark either the one or the other population genetic
process. Especially for addressing adaptively relevant DNA polymorphism,
sequence analysis of the relevant DNA fragments may be useful.
Acknowledgements
Technical assistance by Katrin Groppe is gratefully appreciated, mainly
the procurement of the oak seedlings. We would like to express our gratitude
to Dr. Frank Hartung (IPK, Gatersleben, Germany). He gave helpful advice
on experimental steps in AFLP analysis. Financial support is highly appreciated:
EU, project 'Tree Biodiversity' CT 96 0703 to B.Z., Federal Ministry of
Food, Agriculture and Forestry, Germany, project: 'Biodiversität in
Wäldern' to B.D.
References
Brettschneider R (1996) Non-radioactive AFLP method, based on Digoxigenine. Molecular Screening News 9: 13-14.
Castiglione P, Ajmone-Marsan P, van Wijk R, Motto M (1999) AFLP markers in a molecular linkage map of maize: co-dominant scoring and linkage group distribution. Theoretical and Applied Genetics 99: 425-431.
Cervera MT, Gusmao J, Steenackers M, Peleman J, Storme V, Vanden Broeck A, van Montagu M, Boerjan W (1996) Identification of AFLP molecular markers for resistance against Melampsora larici-populina in Populus. Theoretical nd Applied Genetics 93: 733-737.
Debener T, Mattiesch L (1999) Construction of a genetic linkage map for roses using RAPD and AFLP markers. Theoretical and Applied Genetics 99: 891-899.
Degen B, Ziegenhagen B, Gillet E, Scholz F (1995) Computer-aided search for codominant markers in complex DNA banding patterns - A case study in Abies alba Mill. Silvae Genetica 44: 274-282.
Degen B, Gregorius H-R, Scholz F (1996) ECO-GENE, a model for simulation studies on the spatial and temporal dynamics of genetic structures of tree populations. Silvae Genetica 45: 323-329.
Degen B (1996) CoTrix, Version 3.0/1999 - A computer programme for analysis of DNA banding patterns including user's manual. Internet-publication: http://kourou.cirad.fr/genetique/
Degen B, Caron H, Bandou E et al. (1999) Small scale spatial genetic structure of six tropical tree species in French Guiana, submitted paper.
Deichsel G, Trampisch HJ (1985) Clusteranalyse und Diskriminanzanalyse. Gustav Fischer Verlag, Stuttgart.
Dow BD, Ashley MV (1996) Microsatellite analysis of seed dispersal and parentage of saplings in bur oak. Quercus macrocarpa. Molecular Ecology 5: 615-627.
Dumolin S, Demesure B, Petit RJ (1995) Inheritance of chloroplast and mitochondrial genomes in pedunculate oak investigated with an efficient PCR method. Theoretical and Applied Genetics 91: 1253-1256.
de Greef B, Triest L, de Cuyper B, van Slyckens J (1998) Assessment of intraspecific variation in half-sibs of Quercus petraea (Matt.) Liebl. 'plus' trees. Heredity 81: 284-290.
Gerber S, Mariette S, Streiff R, Bodénès C, Kremer A (1999) Comparison of microsatellites and AFLP markers for parentage analysis. Chapter 4 in this Compendium: Gillet, E.M. (ed.). Which DNA Marker for Which Purpose? Final Compendium of the Research Project Development, optimisation and validation of molecular tools for assessment of biodiversity in forest trees in the European Union DGXII Biotechnology FW IV Research Programme Molecular Tools for Biodiversity. URL http://webdoc.sub.gwdg.de/ebook/y/1999/whichmarker/index.htm
Jones CJ, Edwards KJ, Karp A et al. (1998) Reproducibility testing of AFLPs by a network of European laboratories. In: Karp A, Isaac PG, Ingram DS (eds.). Molecular Tools for Screening Biodiversity. Chapman and Hall, London, pp. 191-192.
Lexer C, Heinze B, Steinkellner H, Kampfer S, Ziegenhagen B, Glössl J (1999) Microsatellite analysis of maternal half-sib families of Quercus robur (pedunculate oak): detecting seed contaminations and inferring the seed parents from the offspring. Theoretical and Applied Genetics 99: 185-191.
Lexer C, Streiff R, Steinkellner H, Glössl J (1997) Vaterschaftstests für Bäume mit Mikrosatelliten. Österreichische Forstzeitung 108: 43-44.
Lexer C, Heinze B, Gerber S, Steinkellner H, Ziegenhagen B, Kremer A, Glössl J (1999) Chapter 6 in this Compendium: Gillet, E.M. (ed.). Which DNA Marker for Which Purpose? Final Compendium of the Research Project Development, optimisation and validation of molecular tools for assessment of biodiversity in forest trees in the European Union DGXII Biotechnology FW IV Research Programme Molecular Tools for Biodiversity. URL http://webdoc.sub.gwdg.de/ebook/y/1999/whichmarker/index.htm
Lin J-J, Ambrose M, Kuo J (1997) Chemiluminescent detection of AFLPTM fingerprints. Focus 19: 36-38.
Pakniyat H, Powell W, Baird LL et al. (1997) AFLP variation in wild barley (Hordeum spontaneum C. Koch) with reference to salt tolerance and associated ecogeography. Genome 40: 332-341.
Paul S, Wachira FN, Powell W, Waugh R (1997) Diversity and genetic differentiation among populations of Indian and Kenyan tea (Camilla sinensis (L.) O. Kuntze). Theoretical and Applied Genetics 94: 255-263.
Prabhu RR, Gresshoff PM (1994) Inheritance of polymorphic markers as generated by DNA amplification fingerprinting and their use as genetic markers in soybean. Plant Molecular Biology 26: 105-116.
Robinson JP, Harris SA (1999) Amplified Fragment Length Polymorphisms and Microsatellites: A phylogenetic perspective. Chapter 12 in this Compendium: Gillet, E.M. (ed.). Which DNA Marker for Which Purpose? Final Compendium of the Research Project Development, optimisation and validation of molecular tools for assessment of biodiversity in forest trees in the European Union DGXII Biotechnology FW IV Research Programme Molecular Tools for Biodiversity. URL http://webdoc.sub.gwdg.de/ebook/y/1999/whichmarker/index.htm
Steinkellner H, Fluch S, Turetschek E, Lexer C, Streiff R, Kremer A, Burg K, Glössl J (1997) Identification and characterization of (GA/CT)n- microsatellite loci from Quercus petraea. Plant Molecular Biology 33: 1093-1096.
Streiff R, Labbe T, Bacilieri R, Steinkellner H, Glössl J, Kremer A (1998) Within-population genetic structure in Quercus robur L. and Quercus petraea (Matt.) Liebl. assessed with isozymes and microsatellites. Molecular Ecology 7: 317-328.
Streiff R, Ducousso A, Lexer C, Steinkellner H, Glössl J, Kremer A (1999) Pollen dispersal inferred from paternity analysis in a mixed oak stand of Quercus robur L. and Quercus petraea (Matt.) Liebl. Molecular Ecology 8: 831-841.
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Mornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research 23: 4407-4414.
Vrieling K, Peters J, Sandbrink H (1997) Amplified Fragment Length Polymorphisms (AFLPs) detected with non-radioactive digoxigenine labelled primers in three plant species. Plant Molecular Biology Reporter 15: 255-262.
Ziegenhagen B, Guillemaut P, Scholz F (1993) A procedure for mini-preparation of genomic DNA from needle tissue of silver fir (Abies alba Mill.). Plant Molecular Biology Reporter 24 (2): 117-121.
Ziegenhagen B, Brettschneider R, Kuhlenkamp V, Fladung M (1999) Non-radioactive labelled AFLPs for application in forest trees, submitted paper.
© Institut für Forstgenetik und Forstpflanzenzüchtung, Universität Göttingen, 1999