D-Lib Magazine
September/October 2016
Volume 22, Number 9/10
Table of Contents
Preliminary Study on the Impact of Literature Curation in a Model Organism Database on Article Citation Rates
Tanya Berardini
The Arabidopsis Information Resource, Redwood City, California
tberardi@arabidopsis.org
Ron Daniel Jr.
Elsevier Labs, Concord, California
r.daniel@elsevier.com
Michael Lauruhn
Elsevier Labs, Portland, Oregon
m.lauruhn@elsevier.com
Leonore Reiser
The Arabidopsis Information Resource, Redwood City, California
lreiser@arabidopsis.org
DOI: 10.1045/september2016-berardini
Printer-friendly Version
Abstract
Literature curation by model organism databases (MODs) results in the interconnection of papers, genes, gene functions, and other experimentally supported biological information, and aims to make research data more discoverable and accessible to the research community. That said, there is a paucity of quantitative data about if and how literature curation affects access and reuse of the curated data. One potential measure of data reuse is the citation rate of the article used in curation. If articles and their corresponding data are easier to find, then we might expect that curated articles would exhibit different citation profiles when compared to articles that are not curated. That is, what are the effects of having scholarly articles curated by MODs on their citation rates? To address this question we have been comparing the citation behavior of different groups of articles and asking the following questions: (1) given a collection of 'similar' articles about Arabidopsis, is there a difference in the citation numbers between articles that have been curated in TAIR (The Arabidopsis Information Resource) and ones that have not, (2) for articles annotated in TAIR, is there a difference in the citation behavior before vs. after curation and, (3) is there a difference in citation behavior between Arabidopsis articles added to TAIR's database and those that are not in TAIR? Our data indicate that curated articles do have a different citation profile than non-curated articles that appears to result from increased visibility in TAIR. We believe data of this type could be used to quantify the impact of literature curation on data reuse and may also be useful for MODs and funders seeking incentives for community literature curation. This project is a research partnership between TAIR and Elsevier Labs.
Keywords: Model Organism Databases (MODs); Impact Factor; Citation Analysis
1 Introduction
In the era of big data, curated databases are an essential research tool yet little is known about the impacts of scientific literature curation by biological databases and how that activity affects use and reuse of data from the primary literature. Many model organism databases (MODs) curate the primary literature, a process in which experimental data is extracted and associated to database objects and rendered into machine readable formats. Literature annotation is a time consuming and costly process that is done manually by professional curators with domain expertise in their organism. While the goal in doing all this work is to increase data accessibility, discoverability and reuse, we have few ways to measure this outcome. In this study, we attempted to identify means by which we can assess the impact of one aspect of literature curation, functional annotation. Using citation number as a proxy, we sought to address the question "What effect does functional annotation from the literature have on citation rates for publications used for annotation?" If there is a significant impact then it might induce publishers and authors to partner with MODs to include and even require gene function annotations as part of the publication process.
1.1 TAIR literature curation and workflow
TAIR (The Arabidopsis Information Resource) is a curated genome database for Arabidopsis thaliana, an annual weed commonly known as mouse ear cress. TAIR presents a comprehensive overview of the function of each of the plant's more than 30,000 genes and, among other services, provides access to experimental data generated by the research community (Huala, et al., 2001). These data include an extensive corpus of scientific literature that spans almost a century. TAIR has been curating this literature for nearly 15 years: curation includes indexing citations and abstracts in the database, linking articles to relevant data objects (e.g. genes, keywords, mutations) and encoding experimental data in the form of structured associations using controlled vocabularies, termed annotations, that are linked to both data objects and research articles (Berardini, et al., 2004; Li, et al., 2012). In addition to annotation by TAIR curators, members of the research community also annotate articles (Berardini, 2012). Generally it is the authors of those articles that contribute annotations to TAIR although any registered community member is welcome to submit data. TAIR annotations include Gene Ontology (GO) annotations and Plant Ontology (PO). TAIR's annotations are then further distributed via sites such as the Gene Ontology Consortium (Gene Ontology Consortium, 2013), the National Center for Bioinformatics Resources (NCBI), RefSeq, Araport (Krishnakumar, et al., 2015) and others.
Figure 1 summarizes the primary workflow for processing literature for curation in TAIR. Each month TAIR downloads all PubMed citations containing the term 'Arabidopsis' in the title, keyword or MESH terms. These articles are then entered into our curation database (Yoo, et al., 2006; Li, et al., 2012) where a text mining algorithm is used to identify potential matches of gene names in the titles and abstracts to Arabidopsis genes in TAIR. The first pass of manual curation distinguishes those papers that actually refer to Arabidopsis genes (valid) from those that do not (invalid). Invalid papers are typically about another organism but may mention Arabidopsis peripherally. Valid articles are then manually reviewed to verify gene name matches. If the article does not contain data that can be used for gene annotation the article is made 'invalid'. Other examples of invalid articles include those that are primarily reporting technical advances or physiological studies. If the article can be used for gene annotation, it remains valid and in the curation queue (valid, unannotated articles). Article curation is not necessarily done in the order of article submission. Instead they are often curated based upon wanting to capture data on a particular gene.
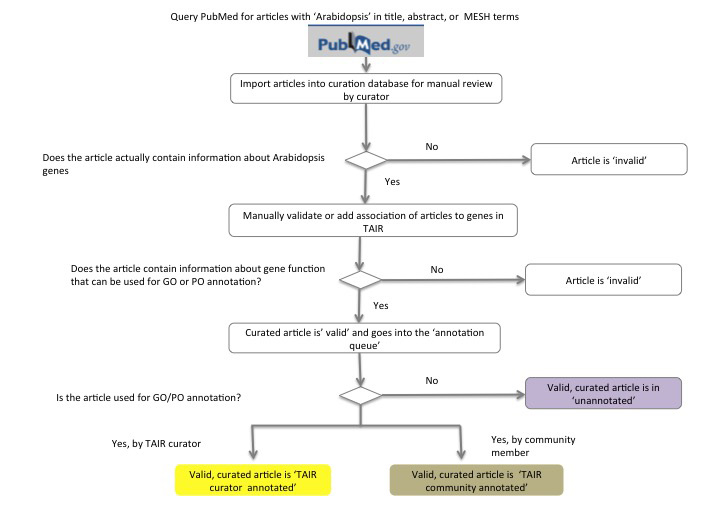
Figure 1: TAIR Literature Curation — Article Classification Workflow
2 Data Collection
To generate our initial dataset we selected articles from TAIR that were either annotated by TAIR curators, annotated by community members or not yet annotated (Figure 1). The raw data included the PubMed ID and for those articles used for annotation, the annotation date, and whether the annotation was done by a community or TAIR staff curator. In some cases articles had more than one curation date and all dates were recorded. We then queried the Scopus bibliographic database with this list of PubMed IDs and obtained the following data for each article: Scopus ID number, Journal ISSN, Source Title, Article Title, Publication Date, Annotation Date (TAIR and Author annotated; Figure 2).
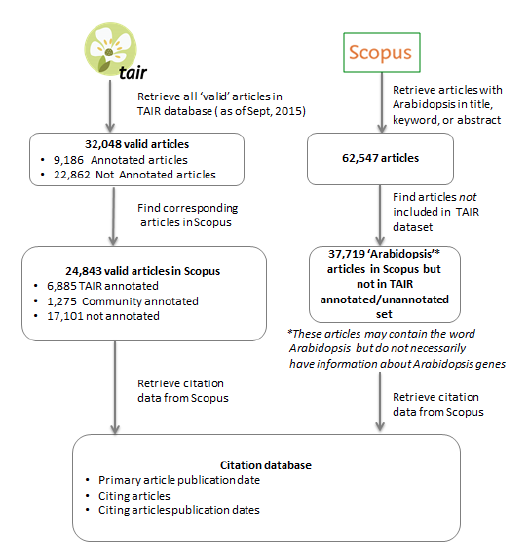
Figure 2: Generating a Database of Articles and Their Citations for Analysis
We then grouped the articles into categories (Table 1, sets A, B and C) according to their annotation status in TAIR. We then further subdivided the data sets based on the journals in which the publications appeared. One set represented the major plant biology journals (The Plant Cell, Plant Physiology, and The Plant Journal) where a large fraction of the total annotated publications are published. The other set represents all other journals.
| Article Set |
Composition |
Number of Papers |
| The Plant Journal, The Plant Cell, Plant Physiology |
| Set A |
TAIR Annotated |
3,034 |
| Set B |
Author + Community Annotated |
847 |
| Set C |
Unannotated |
3,987 |
| Set D |
"Arabidopsis" articles in Scopus that are not in A, B or C |
3,311 |
| All Other Journals |
| Set A |
TAIR Annotated |
3,851 |
| Set B |
Author + Community Annotated |
428 |
| Set C |
Unannotated |
13,024 |
| Set D |
"Arabidopsis" articles in Scopus that are not in A, B or C. |
34,408 |
Table 1: Article Data Sets. Raw data for sets A-C were retrieved from TAIR (10/2015). Data set D was obtained from Scopus (retrieved 10/2015). Citations for all datasets were retrieved from Scopus 10/2015. The total number of articles in A, B and C is greater than the total set of TAIR supplied articles because the same article can be annotated by both TAIR curators and community.
Using the resulting Scopus IDs for the primary articles, we then re-queried the Scopus database and obtained information about the articles that cited the original articles. This query generated a list that contained the original Scopus ID, the Scopus ID for the citing article, the Publication date (in yyyymm format), and the citing article title. At the time this report was generated, the Scopus database contained 24,834 unique articles that TAIR had identified that had been cited a total of 1,568,301 times.
We also collected citation behavior for 'Arabidopsis' articles that were not considered 'annotatable' by TAIR (i.e. did not contain experimental gene function data about Arabidopsis genes). We queried Scopus for articles with Arabidopsis in the Title, Abstract, or Keyword. This returned a total of 62,547 articles. Of those, a total of 37,719 (Table 1, combined set D) were not included in any of the three TAIR datasets (sets A, B and C). This set includes papers that may or may not be about Arabidopsis and may or may not describe gene functions. This would, for example, include methods, experimental information about gene function for non-Arabidopsis genes, 'omics' papers, or physiological studies.
3 Results and Discussion
3.1 Citation rates of selected article sets
After applying filters to the various collections of articles, we retrieved the total number of times the collections were cited and calculated the median number of times each the articles were cited (Figure 3).
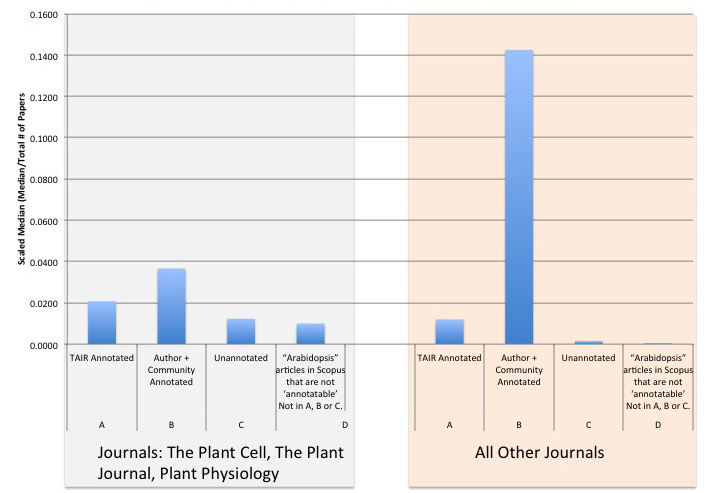
Figure 3: Scaled Median of Citation Counts per Article Type Category
To account for differences in the sizes of the data sets for each category, we also calculated the scaled median citation number, that is the median divided by the total number of papers in each category. The collections that were most cited were those articles used for gene annotation (Table 1, sets A and B). Articles in TAIR database that were used for GO or PO annotations were cited on average 83 times, with a median of 51 citations (data not shown). As mentioned in the previous section, this collection was subdivided into articles annotated by TAIR staff and articles annotated by the article authors or the community. The articles annotated by authors or the community (set B) had higher citation numbers (scaled median of 0.0366 for major plant journals and 0.1425 for all other journals) while the TAIR annotated articles (set A) had a lower scaled median (0.0208 for major plant journals and 0.0119 for all other journals). Articles that were linked to genes in TAIR but not yet used for annotation (set C, not yet annotated) had citation numbers lower than both the staff and author curated articles (scaled median of 0.0123 for major plant journals and 0.0015 for all other journals ). Set D represents the total set of articles that were retrieved from Scopus via the Title, Abstract, and Keyword search (62,547 articles) minus the articles that comprise sets A, B, and C. Set D had a scaled median citation of 0.01 for major plant journals and 0.0001 for all other journals.
3.2 Selection of control groups and collection bias
Preliminary data suggests that articles in TAIR that are used for annotation, especially if they are annotated by the authors themselves or the community, are more likely to be cited than those that are not used for annotation. Both TAIR curator annotated (set A) and community annotated articles (set B) showed an increase in the number of citations when compared to similar types of articles that were not yet annotated (set C). Overall, articles used for annotation (A and B) or that could be used for annotation (set C) were more likely to be cited than those in set D. While the data show that articles used for annotations have more citations than those that are not annotated, it is not possible to state conclusively that there is a direct correlation between annotation and citation rates. This is due to the many challenges in collecting data for this experiment, identifying control data sets, and selection bias.
Creating a control group of articles presents a series of challenges. We selected the unannotated (set C) articles as being most similar to the annotated articles in terms of content and sources. This side-by-side comparison shows that annotated articles in the TAIR database have more citations than unannotated articles. However, we cannot rule out that the effect may be due to one or more types of selection bias due to 'cherry picking' of articles to annotate. TAIR has a partnership with about 17 publishers, some of which provide TAIR with lists of newly published articles and the corresponding author's contact information (Berardini, 2012). Therefore the author annotated article set is predominantly from high profile plant journals. We attempted to look at this potential source of bias by splitting the data sets into publications in the major plant journals and those in all other journals (Figure 3).
As another control we also looked at articles that were not selected for annotation (set D). Articles annotated by TAIR have nearly four times the citations as those in set D. There are many possible explanations for this. Set D includes articles that may mention Arabidopsis genes only peripherally but mostly present information about other species, or if they are about Arabidopsis they may be reviews, describe physiological studies, techniques or large scale studies not captured by TAIR's literature curation pipeline. There may also be an effect based on research area; a paper describing novel gene function in Cacao may simply be less widely read and cited than a similar paper about an Arabidopsis gene.
3.3 Sources of selection bias and future directions
One of the challenges in performing this type of citation analysis is to identify and control for biases that may skew results (Niyazov, 2016). TAIR curators typically select articles that are often the first published description of a gene's function and therefore present novel findings. This may result in a bias towards more noteworthy research that is already highly cited. We still want to delve into effects of time from publication vs. time from annotation on citation numbers to see if it is possible to identify a temporal pattern to increases in citation rate for curated papers. Future studies will need to account for and address temporal factors as well sources of bias such as open access vs. paid articles, author self-selection, research area selection, relative distributions of journals in the sets and journal impact factor. The preliminary results presented here are a first insight and a positive step towards having a metric that can be used to quantify the effects of literature curation, and the efforts of the biocuration community, on the progress of research. While this study represents data from one MOD, TAIR, studies with other MODs are in the planning stages and will provide even more data to examine this important question.
Acknowledgements
The authors would like to thank Barbara Warsavage for advice and guidance on statistics. We would also like to thank members of Elsevier Labs, the Elsevier Scopus team, and Mike Appel for technical assistance.
References
| [1] |
Berardini TZ, Li D, Muller R, Chetty R, Ploetz L, Singh S, Wensel A, Huala E. 2012. Assessment of community-submitted ontology annotations from a novel database-journal partnership. Database. http://doi.org/10.1093/database/bas030 |
| [2] |
Berardini TZ, Mundodi S, Reiser L, Huala E, Garcia-Hernandez M, Zhang P, Mueller LA, Yoon J, Doyle A, Lander G, Moseyko N, Yoo D, Xu I, Zoeckler B, Montoya M, Miller N, Weems D, Rhee SY. 2004. Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiology 135:745-755. http://doi.org/10.1104/pp.104.040071 |
| [3] |
Gene Ontology Consortium. 2013. Gene Ontology annotations and resources. Nucleic Acids Research. Volume 41 (Database issue), Issue D-1, pp D530-5. http://doi.org/10.1093/nar/gks1050 |
| [4] |
Huala E, Dickerman AW, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, Hanley D, Kiphart D, Zhuang M, Huang W, Mueller LA, Bhattacharyya D, Bhaya D, Sobral BW, Beavis W, Meinke DW, Town CD, Somerville C, Rhee SY. 2001. The Arabidopsis Information Resource (TAIR): A comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Research 29:102-105. |
| [5] |
Krishnakumar V, Hanlon MR, Contrino S, Ferlanti ES, Karamycheva S, Kim M, Rosen BD, Cheng CY, Moreira W, Mock SA, Stubbs J, Sullivan JM, Krampis K, Miller JR, Micklem G, Vaughn M, Town CD. Araport: the Arabidopsis information portal. Nucleic Acids Research. 2015. Volume 43 (Database issue), Issue D1, pp. D1003-9. Epub 2014 Nov 20. http://doi.org/10.1093/nar/gku1200 |
| [6] |
Li D, Berardini TZ, Muller RJ, Huala E. 2012. Building an efficient curation workflow for the Arabidopsis literature corpus. Database 2012:bas047. http://doi.org/10.1093/database/bas047 |
| [7] |
Niyazov Y, Vogel C, Price R, et al. Open Access Meets Discoverability: Citations to Articles Posted to Academia.edu. Dorta-González P, ed. PLoS ONE. 2016;11(2):e0148257. http://doi.org/10.1371/journal.pone.0148257 |
| [8] |
Yoo D, Xu I, Berardini TZ, Rhee SY, Narayanasamy V, Twigger S. PubSearch and PubFetch: a simple management system for semiautomated retrieval and annotation of biological information from the literature. Current Protocols in Bioinformatics. 2006, March, Chapter 9, Unit 9.7. http://doi.org/10.1002/0471250953.bi0907s13 |
About the Authors
Tanya Berardini is a Scientific Curator at The Arabidopsis Information Resource and Phoenix Bioinformatics. She has been involved in ontology development, literature curation, data integration, and involving the research community in data contributions for almost 15 years. She obtained her Ph.D in Biology from Boston University.
Ron Daniel Jr. is the Director of Elsevier Labs, an R&D group which concentrates on the future of scholarly communications. Educated as an electrical engineer, Ron has done extensive work on metadata standards such as the Dublin Core, RDF, and PRISM. Before joining Elsevier five years ago, he worked at a startup that was acquired for its automatic classification technology, and consulted on taxonomy and information management issues for nine years. Ron received his Ph.D. in Electrical Engineering from Oklahoma State University, and was a postdoctoral researcher at Cambridge University and at Los Alamos National Laboratory.
Michael Lauruhn is a librarian working in research areas including Linked Data, taxonomies and ontologies, mark-up and annotation, research data lifecycles, and other issues affecting research communications. Before joining Labs in 2010, he held consulting and technical positions helping large companies and organizations define and implement taxonomies and metadata schemas. He received his M.L.S from UCLA.
Leonore Reiser is a Scientific Curator for The Arabidopsis Information Resource and Phoenix Bioinformatics where she curates research literature about the model plant Arabidopsis thaliana and helps develop tools for TAIR. Previously she managed programs aimed at diversifying the sciences. She obtained a Ph.D. in Plant Biology from the University of California.