D-Lib Magazine
January/February 2017
Volume 23, Number 1/2
Table of Contents
HyWare: a HYbrid Workflow lAnguage for Research E-infrastructures
Leonardo Candela
NeMIS Lab, ISTI CNR, Pisa Italy
leonardo.candela@isti.cnr.it
Fosca Giannotti
KDD Lab, ISTI CNR, Pisa Italy
fosca.gianotti@isti.cnr.it
Valerio Grossi
KDD Lab, ISTI CNR, Pisa Italy
valerio.grossi@isti.cnr.it
Paolo Manghi
NeMIS Lab, ISTI CNR, Pisa Italy
paolo.manghi@isti.cnr.it
Roberto Trasarti
KDD Lab, ISTI CNR, Pisa Italy
roberto.trasarti@isti.cnr.it
Corresponding Author: Paolo Manghi, paolo.manghi@isti.cnr.it
https://doi.org/10.1045/january2017-candela
Abstract
Research e-infrastructures are "systems of systems", patchworks of tools, services and data sources, evolving over time to address the needs of the scientific process. Accordingly, in such environments, researchers implement their scientific processes by means of workflows made of a variety of actions, including for example usage of web services, download and execution of shared software libraries or tools, or local and manual manipulation of data. Although scientists may benefit from sharing their scientific process, the heterogeneity underpinning e-infrastructures hinders their ability to represent, share and eventually reproduce such workflows. This work presents HyWare, a language for representing scientific process in highly-heterogeneous e-infrastructures in terms of so-called hybrid workflows. HyWare lays in between "business process modeling languages", which offer a formal and high-level description of a reasoning, protocol, or procedure, and "workflow execution languages", which enable the fully automated execution of a sequence of computational steps via dedicated engines.
Keywords: HyWare, Workflow Language, Research E-infrastructure
1 Introduction
Over the past decade Europe has developed world-leading expertise in building and operating e-infrastructures.1 They are large scale, federated and distributed research environments in which researchers have shared access to unique scientific facilities (including data, instruments, computing and communications), regardless of their type and location in the world. They are meant to support unprecedented scales of international collaboration in science, both within and across disciplines. Their aim is to realise a common environment where scientists can create, validate, assess, compare, and share their digital results of science, such as research data, intended as scientific data produced by a scientific effort, and research methods, intended as digital computation-oriented elements resulting from their research; research methods are discipline-specific, as well as research data, but examples include software, services, tools, workflows, scripts, algorithms, and protocols.
The digitalization of the scientific process has raised unprecedented opportunities and challenges in the way science can be performed but also shared and reused. In the last decade, all stakeholders of the research life-cycle (e.g. researchers, organizations, funders) have highlighted and endorsed the importance of applying Open Science publishing principles
The digitalization of the scientific process has raised unprecedented opportunities and challenges in the way science can be performed but also shared and reuse. In the last decade, all stakeholders of the research life-cycle (e.g. researchers, organizations, funders) have highlighted and endorsed the importance of applying Open Science publishing principles (Bartling et al., 2014). According to such principles, researchers should "publish" their scientific results in order to enable transparent evaluation and reproducibility of science. According to such vision, the scientific article is only one of the possible publishable products, certainly required but insufficient at supporting Open Science principles. The Open Science movement encourages researchers to "publish" (i) research data and methods valuable to their research (e.g. input and output data, a text-mining algorithm), (ii) the e-infrastructure tools and services they used to implement their research, and possibly (iii) their research workflow, intended as the sequence of research actions they performed to reach their results. Such actions are intended as the steps the researches had to perform using the e-infrastructure tools to run an experiment (e.g., I used tool X over research data Y to test my research method Z). The availability of all the constituent parts and results of a scientific process, which is in turn described in an article, maximizes the chances to correctly evaluate the quality of the research and the chances to re-use results produced by others (reducing the cost of science). In particular, where available, the availability of workflows is of paramount importance in order to address automated execution of scientific experiments and, more generally, to enable reproducibility of science (LeVeque et al., 2012).
The implementation of Open Science principles is hindered by a multitude of problems. One of the most prominent is that e-infrastructures available to research communities today are often far from being well-designed and consistent environments embracing all needs of a research community. They are rarely (if ever) designed from scratch to (i) uniformly share and reuse products of science, be they research datasets and methods, and to (ii) describe the scientific process via workflows made of research actions performed over homogeneous and machine executable e-infrastructure tools, i.e. common policies, standards, language platforms, and combinable APIs. They are rather "systems of systems", patchworks of tools that process or generate research products, and are often equipped with "catalogues" where researchers (and services) can register both tools and products to enable their sharing, discovery and reuse. The heterogeneity of tools is often a feature, not a bug, as it is not possible to assume that scientists will always produce e-infrastructure tools according to a rigorous workflow language-like approach (e.g. Apache Taverna (Oinn et al., 2004)). Indeed, while subsystems of e-infrastructures may support workflow languages and engines, this way of thinking can hardly be imposed on the research e-infrastructure as a whole, namely cross-platform, cross-nation, cross-lab, cross-funders. For example, actions of a workflow may be: invoke a web service A over data d and collect the result d'; download a tool for the execution of R-scripts and execute the script r over the data d'; collect the Taverna workflow t and execute it on myexperiment.org over your data. Accordingly, the scientific process (the sequence of actions performed by the scientists) cannot, in general, be uniformly described, shared, and therefore re-executed. Rather, it is concretized in a narrative description (e.g. scientific article), thereby hindering transparent evaluation and reproducibility of experiments. E-infrastructures are missing the frameworks and tools needed to unambiguously describe such "hybrid" actions, combine them into "hybrid" workflows, share the "hybrid" workflows, and provide machine-support (to the extent made possible by the action typology) to the execution of "hybrid" workflows.
In this paper we present HyWare, a language for the specification of "hybrid" workflows reflecting the scientific process in highly-heterogeneous e-infrastructures. HyWare lays in-between business process modeling languages (Rosemann et al., 2000), which offer a formal and high-level description of a reasoning, protocol, or procedure, and workflow execution languages (e.g. BPEL (Weerawarana et el., 2005)), which enable the fully automated execution of a sequence of computational steps via dedicated engines. HyWare defines a framework where research actions templates (identifying classes of actions) can be customized according to the specifics of the underlying e-infrastructure, and then be combined by researchers into workflows. HyWare-based tools will offer user interfaces to support the scientists when constructing a workflow and make it available for others to discover. A second scientist may later discover the workflow and, via the same HyWare-based tool, be guided through its execution on a step-by-step basis. Such tools display to the researchers which steps they should manually execute to repeat the experiments but, when a sequence of steps is based on components of executable workflows, execute automatically the relative subpart.
The remainder of the paper is organised as follows: Section 2 describes the HyWare language and its constituents; Section 3 describes how the HyWare-based approach can be successfully implemented in the case of SoBigData.eu, a large scale Research Infrastructure intended to serve the Social Mining research community; and Section 4 concludes the paper by reporting on future works.
2 Workflow Language
One of the aims of a research e-infrastructure (e-infra) is to guarantee an integrated framework in order to provide the researchers with a homogeneous and expressive way for representing experiments, performing and sharing them among the community. Consider the following scenario: a scientist working with the e-infra is using its different tools to run the experiments, which consist of reusing and generating research data and methods by means of sequences of actions of the following kinds:
- Local execution of tools to be downloaded and installed: the execution of the action requires the user to download and execute the tool on its own premises;
- Call to web-accessible services (SOAP or REST): the execution of the action requires a call to the service that is operated by a provider;
- Web-accessible applications (tools accessible via user interfaces from the web): the execution of the action requires accessing the web user interface;
- Execution of a workflow: the execution of the action requires invoking the respective workflow execution engine;
- Manual operations: the execution of the action consists of describing the effect of an action rather than the steps to execute it, based on the assumption that other scientists have the required know-how (e.g. make this file accessible from the internet via a URL).
Once the experiment is concluded, the scientist has identified the actions he/she needs to perform to reproduce it and he/she is now willing to materialize the relative workflow in order to share it with others. This will allow other researchers to be convinced of the quality and value of his/her scientific process, by possibly repeating and reproducing the experiment for validation purposes or reusing its parts. To this aim, the scientists need a workflow language capable of describing and combining machine-executable actions and human-executable actions in such a way that the workflow can be re-executed in a trustable and objective way, guaranteeing evaluation and reproducibility of science. This language has to orchestrate actions resulting from tools and best practices made available to the e-infra and interpreted by different scientists, with no common agreements and policies on the way the flow between such actions has to be implemented, e.g. how input and output data flows from one action to another. The literature has focused on two main types of workflow modeling language:
- Human-oriented workflow languages. Such languages are able to conceptually represent a set of steps that drives the user to reproduce the experiments, but does not include any machine execution support to the individual actions, which are to be performed by the scientists. To this aim, generic workflow languages can be adopted, such as UML (UML, 2005), YAWL (Aalst et al., 2005), and JBPM for Business process (jBMP, 2016), but also more specific scientific workflow languages exist, such as the one proposed by protocols.io. The limitation of this class of solutions is that they are fully decoupled from the e-infra and do not interact with its components, even when some of the actions could be automated. For example, with respect to the scenario above, an action could show the instructions on how and where data can be downloaded and what the requirements of the software are, where it can be downloaded and executed, but could not execute a web service on behalf of the user. On the other hand, the benefits of such solutions come from their flexibility and adaptability to all possible e-infra. The highest level of machine support is the one proposed by protocols.io, which offers a virtual environment for scientists of different communities to create and share their "protocols" with a DOI (i.e. a web persistent identifier for scholarly communication resources), intended as workflows of scientific human actions. The application offers the possibility to "run" the protocols, which consists in prompting the scientists with a sequence of tasks (one per action) and the functionality to move from completed tasks to the next to be completed.
- Machine-oriented workflow languages. Such languages are able to specify workflows as a sequence of actions to be executed by an e-infra "interpreter", underlying and strictly coupled with the language. To this aim, the actions must be based on tools conforming to minimal integration requirements imposed by the e-infra middleware. The execution of the workflow is managed by the middleware (e.g. workflow engine), which is responsible for both the computation of the single action and the orchestration of all the actions, including datasets and software movements. The scientists can therefore describe, share, and re-execute experiments encoded as workflows, changing input parameters and re-using parts of the nodes. On the other hand, scientists building tools for the e-infra (for the benefit of the community) are bound to develop these in accordance with the minimal integration requirements of the middleware. This approach works nicely around sub e-infras, namely subsystems and frameworks from which they scientists can benefit on rather specific sub-topics, but in general cannot be applied to the overall e-infra, which may include the usage of third-party cross-disciplinary services, not interested in specific integration with specific e-infras. In (Ceri et al., 2013) a theoretical vision of how a mega-modeling language can integrate different tools and actions is presented, meanwhile several existing systems of this kind of workflow languages are KNIME (Berthold et al., 2007), SAS (Sas, 2011), Meandre (Llora et al., 2008), Kepler (Ludäscher et al., 2006), and Apache Taverna (Oinn et al., 2004).
HyWare falls in the intersection between the two categories above: the orchestration of the workflow is performed by the e-infra, some of the actions may be executed by the e-infra, but other actions are executed by the scientists. HyWare takes inspiration from:
- The language KNIME. KNIME graphically describes a workflow as a directed graph of nodes whose input and output parameters are interconnected and validated according to a degree of compatibility, and whose edges imply a temporal ordering of execution; KNIME workflows are executed by a local engine, which executes their business logic and lets the data flow across them, to produce a final output;
- The protocols.io approach. Actions that require human interaction are described according to given templates, so that other scientists can interpret them and execute them; such actions become KNIME-like nodes and are "executed" by the e-infra in the sense the middleware prompts the scientist with the instructions to perform the action; in accordance with KNIME features, also in HyWare, human actions conform to minimal specification requirements, which allow input and output of the actions to flow from machine executable to human executable nodes.
HyWare hybrid workflows are built out of two main classes of action nodes: (i) the actions that require human interaction, and (ii) the actions that are performed by the e-infra. Unlike other languages, however, the workflow makes sure that research data or research methods flow through both classes of nodes in a coherent way, also subject to compatibility patterns. As in the case of protocols.io, the language does not enforce specific kinds of actions but rather offers the possibility to instantiate e-infra specific classes of human actions (e.g. download and execution of software) or machine actions (e.g. web service call) based on a meta-structure for actions, which includes: (i) the name of the class, (ii) a sequence of descriptive properties whose values describe the specific action instance, (iii) a list of input and output parameters types, and (iv) the type of the class, namely human-actions or machine-actions.
- Human actions. Human actions are characterized by a description, expressed by the respective properties, which should be detailed enough to guide a scientist through the execution of the action. Such actions should reflect the flavor of machine actions, in the sense their performance should cause the execution of software over a machine in order to compute over input parameters. For example, the operation of downloading software and installing it cannot be regarded as a HyWare action. The action of downloading software, installing it, and executing it over input data can instead be considered a valuable HyWare action.
- Machine actions. Machine actions are characterized by a description, expressed by the respective properties, but also by a standard way to invoke a third-party service and get back the results. To this aim, for each machine action class, the HyWare workflow engine must integrate a mediator capable of invoking the external service with given action input parameters and collect the parameters in order to return them in accordance to the action output type.
- Hybrid workflows. A hybrid workflow is a composition of human and/or machine actions into a Directed Acyclic Graph (DAG). Time edges between actions express the chronological ordering of the actions, hence, before an action can begin all actions relative to incoming time edges must be terminated. As in the case of KNIME (and similar languages) between two nodes connected by a time edge other input-output edges exist, indicating the associations between the output parameters and the input parameters of the two actions.
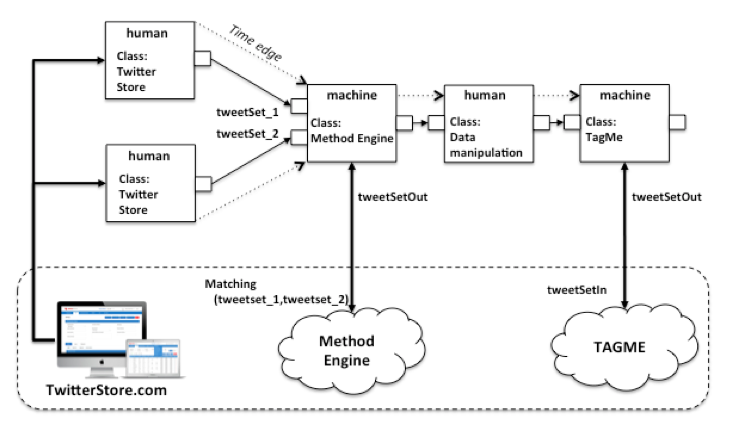
Figure 1: Example of HyWare workflow
For example, consider the workflow in Figure 1, where we assume HyWare has been adopted in an e-infra featuring (i) the web application TwitterStore.com and (ii) two web services Method Engine and TAGME. TwitterStore.com is a web application downloading and collecting sets of tweets based on given tags and time spans over time, which scientists can use to download collections of tweets as CSV files. Method Engine is a web service offering the execution of a set of methods over input data accessible via URLs and returns the URL of the result; one of such methods, i.e. Matching, downloads two sets of tweets 1 and 2 from two URLs, then selects the tweets in 2 only if they are in a relationship R (semantics not important here) with the tweets of 1. TAGME is a web service capable of anchoring Wikipedia entries to words of texts in order to enrich them with correct semantics; the service expects such texts to be provided from a URL in bulk according to a given XML schema. The HyWare workflow engine starts the execution of the workflow from the two actions (that have no input edges) of class Twitter Store. These include a standard description for interacting with such an application which is visualized to the scientists; the instance of such actions can also include the parameters the scientists should use to repeat a given workflow instantiation, i.e. to download specific sets of tweets. Once both sets of tweets have been downloaded on the scientist's desktop, the next action can be executed. As specified by its input types, the action of type Method Engine requires the URLs of the two CSV files. The HyWare engine is aware of the output type of Twitter Store actions (i.e. a local CSV file), hence it (i) instructs the scientists that the preparatory action of uploading the two files on the Web (e.g. on DropBox) must be performed before the action can be effectively executed, and (ii) prompts the user with the request for the two URLs. When the URLs are provided the engine executes the action by invoking the underlying Method Engine web service, which in turn returns the URL of the result. As a completion of the action, the HyWare engine visualizes the URL and moves to the next human action of class Data Manipulation, which consists of a narrative description of what the scientist needs to do to prepare the data for the next machine action: "download the resulting CSV from the URL and prepare an XML file that contains the text of the tweets according to the XML schema available at <URL>". The output type of this action is a local file of the type required by the last machine action of class TAGME. Again, by confronting output and input type, the HyWare engine (i) tells the scientist that the XML file should be uploaded in the web and be accessible from an URL; and (ii), given the URL, fires the TAGME action.
The novelty of HyWare is the ability to cover human and machine actions and to integrate them into the same hybrid workflows at minimal (mandatory) e-infra integration costs. As a result, scientists can describe their scientific process in terms of concretely sharable and reusable workflows, exploiting the best of the autonomic functionalities of the e-infra while respecting the hectic and heterogeneous behavior of scientists in the production of their e-infra tools.
3 SoBigData case study
In this section we present the real case study of the SoBigData.eu e-infrastructure. SoBigData.eu is an European Commission project whose goal is to create the Social Mining & Big Data Ecosystem, i.e. a research infrastructure (RI) providing an integrated ecosystem for ethic-sensitive scientific discoveries and advanced applications of social data mining on the various dimensions of social life, as recorded by "big data". SoBigData.eu is opening up new research avenues in multiple research fields, including mathematics, ICT, and human, social and economic sciences, by enabling easy comparison, reuse and integration of state-of-the-art big social data, methods, and services, into new research. Although SoBigData.eu is primarily aimed at serving the needs of researchers, the openly available datasets and open source methods and services provided by the new research infrastructure will also impact industrial and other stakeholders (e.g. government bodies, non-profit organizations, funders, policy makers).
SoBigData aims at realizing a homogenous e-infrastructure by "gluing together" tools that scientists and practitioners have been realizing in full autonomy and without relying on common interoperability agreements. To this aim, SoBigData e-infrastructure is being realized following an "open ecosystem approach" using the e-infrastructure platform D4science.org as pivot (Assante et al., 2016, Candela et al., 2014). The D4Science platform supports an advanced notion of Virtual Research Environments (VREs), intended as innovative, web-based, community-oriented, comprehensive, flexible, and secure working environments conceived to serve the needs of nowaday scientific investigations (Candela et al., 2013). The implementation and operation of such challenging and evolving working environments largely benefits from, and complements, the offering of Research Infrastructures. Via the pivotal infrastructure it is possible to integrate heterogeneous community tools and offer them to scientists in dedicated VREs, each tailored to satisfy the needs of a designated community. Clearly this represents a challenge, with the pre-requisites described in previous sections: absence of common standards and protocols, variety in technologies and usage modes, inability or unwillingness to change technology. D4Science acts as the technological bridge via which such tools can be registered, discovered and used by scientists, respecting the intent or possibility for the owners of the tools to integrate them fully (e.g. autonomic execution), partly (e.g. web interface integration), or not integrate them in the e-infrastructure (e.g. download and install). In addition, scientists can also benefit from advanced collaborative services: (i) a VRE workspace, organized as a shared file system, which allows data to be moved between different tool-based actions; (ii) a data analytics platform benefitting from a distributed and multi-tenant computing infrastructure oriented to provide scientists a broad variety of algorithms and methods as well as other kind of web-based resources; and (iii) collaboration oriented facilities enabling scientists to publish research results with the possibility to add comments on them in a social-network fashion.
SoBigData scientists can today integrate their tools for VRE-integrated reuse, but cannot represent a sequence of actions as a workflow, in order to share it and reproduce it. Equipping SoBigData VREs with HyWare allows scientists to attach to a specific result the entire process used to obtain it. This makes the environment evolve into a living laboratory, which contains not only the methods and the results but also the experience of the researcher using the methods, and composing an analytical process with it.
In the following sections we shall introduce the classes of actions characterizing SoBigData tools and showcase the usage of HyWare to represent the implementation of an analytical workflow called City of Citizens using HyWare language.
3.1 SoBigData tools and HyWare actions
In this section we describe the SoBigData.eu infrastructure action classes and instances of such classes, i.e. actions, as described and exemplified in Section 2. Moreover, we shall describe how they are made available in the VREs. As described above, the VRE is a working environment tailored to serve the needs of a specific research scenario. The underlying D4Science e-infrastructure platform allows SoBigData scientists to (i) integrate and then register resources, i.e. tools and products (e.g. datasets), to the SoBigData infrastructure and (ii) build VREs as sets of such resources, to support the specific needs of a group of scientists. Specifically, tool resources comprehend and entail the following classes of HyWare actions:
- Method invocation: methods (e.g. Java methods, R algorithms, Python methods) to be executed by the D4Science processing engine, integrated based on OGC Web Processing Service (WPS);
- Software to be downloaded and executed locally;
- Web (REST) services for remote invocation;
- Web applications, offering WebUI-accessible functionalities.
Of course, a general class of manual operation is included. Table 1 reports a sample set of actions currently available to SoBigData.org, together with the relative name, input parameters, output parameters, and actions classes.
Table 1: Example of City of Citizens operators
| Name |
Inputs |
Outputs |
Parameters |
Action class |
| Trajectory Builder |
1-Table (int, double, double, timestamp) |
1-Table (int, geometry) |
Max_time_gap Min_space_gap |
Method |
| Mobility Profiling |
1-Table (int, geometry) |
1-Table (int, profile) |
Maximum_distance Minimum_size |
Software |
| Trajectory Prediction Evaluation |
1-Table (int, geometry double)
2-Table (int, geometry) |
— |
— |
Manual operation |
| Data Splitter |
1-Table |
1-Table
2-Table |
Method Training/Test ratio |
Method |
| Trajectory cutting |
1-Table (int, geometry) |
1-Table (int, geometry) |
— |
Manual operation |
| MyWay |
1-Table (int, profile)
2-Table (id, geometry) |
1-Table (int, geometry double) |
Spatial_tolerance |
Method |
| Twitter Scraper |
— |
1-Json |
Query |
Web Service |
| Network builder |
1-CSV |
1-CSV |
— |
Manual operation |
| Statistical Validation |
1-CSV |
1-CSV |
Noise_percentage |
Method |
| Free Time Computation |
1-CSV |
— |
— |
Software |
For example the Trajectory Builder operator is a Java method completely integrated in the D4Science platform which is able to execute it autonomously. Mobility Profiling is a software library that is available for download and should be executed by the user on a trajectory database on her/his desktop. Finally, Twitter Scraper offers a REST API to download the JSON files representing the tweets with characteristics (e.g. geo-localized tweets in a specific area) required by the query. This mapping between tools of the e-infrastructure and the relative actions allows the user to use HyWare inside the VREs to represent the analytical processes they perform in terms of hybrid workflows, i.e. DAGs of actions. Such workflows are themselves SoBigData product resources, hence sharable and reproducible, by other scientists, as well as subject to comments and discussion. In the next section we present an example of this case in the context of mobility data analysis.
3.2 HyWare workflow for the analytical process of City of Citizens
SoBigData.org has published a number of real-case experiences each grouping analytical processes relevant to a class of problems in social mining. In this section we focus on the experience "City of Citizens" and two of its analytical processes, whose objective is to generate a set of statistics and models describing a territory by means of data, statistics and models. The examples we show are real-case workflows used by the data analyst for (i) testing the MyWay trajectory prediction algorithm (Trasarti et al., 2015), and (ii) analyzing the "usage" of a city by computing people's "free time" (Andrienko et al., 2015).
(1) MyWay Evaluation. In Figure 2 the first example is depicted. Here the process starts with the Trajectory Builder which requires a reference to a table in the platform database containing the spatio-temporal observations of the user in the form: userid, the longitude, latitude and the timestamps. Since this input is not configured, the HyWare interpreter will suggest to the user to enter a URL to locate the table, or how to upload the data to the platform (e.g., how to create a database instance and upload a CSV file into a table) in order to start the workflow. Subsequently, the platform will automatically execute the Trajectory Builder action constructing the trips of the users according to the parameters specified — breaking the sequence of points when the user stops in a place for more than max_time_gap seconds remaining in a spatial buffer of min_space_gap meters. The resulting data is passed to the Data splitter that consists of the invocation of a method capable of dividing the content of a table into a random splitting that respects the training/test ratio, e.g. 60%/40%. The two tables are then used respectively to build the profiles and to test the algorithm. In particular Mobility profiling is a tool available to download and the HyWare interpreter will guide the scientists as to its installation and usage to obtain the result by processing the input table. Trajectory cutting is a manual operation action (no tool is available to the e-infrastructure to perform this action). As such it instructs the scientists on how this task should be accomplished: "taking only a portion of the trajectories by cutting the final part, i.e. removing the last 33% or 66% of the points starting from the end". Since both of the actions are executed local to the scientists desktop, the HyWare interpreter will suggest to the analyst how to upload the results in the platform in such a way that the next action can be executed. The MyWay action is invoked by the scientists to build a predictor from the two input profiles and perform the prediction on the cut trajectories producing a set of predicted trajectories with a score of confidence. Finally, the last human-oriented action will describe how the user should compare the predicted trajectories with the original one (in the test set) in order to perform a Trajectory prediction evaluation.
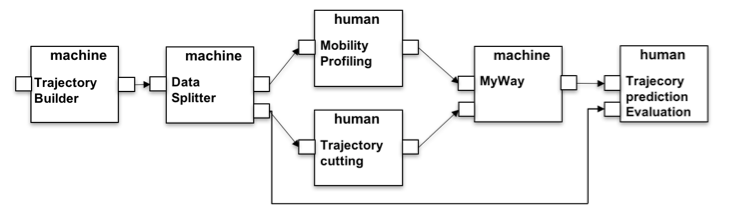
Figure 2: MyWay performance evaluation implemented HyWare workflow
An example of the evaluation test on MyWay is depicted in Figure 3 where different runs of the workflow are represented (using 33% and 66% as parameter for the cutting).
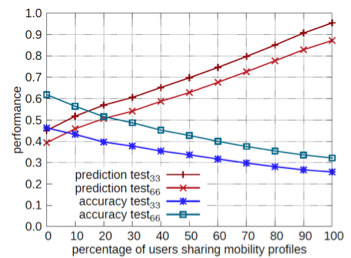
Figure 3: MyWay performance evaluation implemented HyWare workflow
It is interesting to note that removing a smaller amount of points (33% vs 66%) leads to better prediction due the fact that the predictor has more information about the "actual" trip.
(2) Free Time analysis. The second example is shown in Figure 4. The initial action recovers the data from Twitter. The platform itself does not provide a way to do it but the Twitter Scraper action explains (i) how to get a JSON file containing the tweets using the on-line REST API; and (ii) what set of tweets should be downloaded, i.e. geo-localized tweets covering a city area. The former description is part of all instances of the action, which instead will vary depending on the latter parameters. Once the JSON file is obtained, it can be used to build a network according a specific methodology described in the Network building action. In this case the input of the Network building action, i.e. a CSV file, and the output of Twitter Scraper, i.e. a JSON file, do not match; the HyWare interpreter, which can statically detect the difference, can suggest to the user how to perform the type conversion. The network resulting from Network building is then uploaded to the platform workspace which can execute Statistical validation to extract a subnetwork representing the backbone of the original one (performing a statistical check on the arc probabilities). Finally, the result is used in the platform called Common GIS available for download which includes a tool to analyze how the people use specific areas of the city considering their activities extrapolated from the network in input.
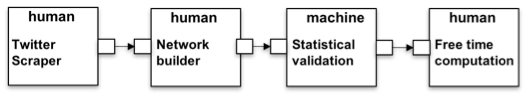
Figure 4: Free Time analysis implemented HyWare workflow
The result is presented visually on the tool and an example is shown in Figure 5, where different areas of the city of Pisa are classified with a different purpose such as:
shopping+leasure,
transport, or
health care.
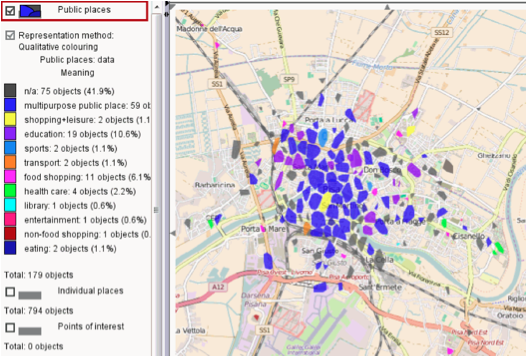
Figure 5: Example of Free Time analysis result
These examples prove the ability of HyWare to represent processes performed by the analysts, where processes are integrating human and machine executable actions to flexibly support a degree of reproducibility of the results in highly heterogeneous scenarios.
4 Future Work
In this paper we presented HyWare, a language for the representation and reproducibility of workflows in highly heterogeneous e-infrastructures. HyWare will be the scientific process workflow language of the SoBigData e-infrastructure, and will cover all the different aspects related to the development of an e-infra for social mining in the context of big data. Based on this vision, the idea is to: (i) complete the formal description of the language by expanding the SoBigData use-cases, and (ii) define a digital encoding of the language in order to support the creation of WebUIs for the construction and execution of workflows.
Acknowledgements
This work is supported by the European Community's H2020 Program under the scheme 'INFRAIA-1-2014-2015: Research Infrastructures', grant agreement #654024 'SoBigData: Social Mining & Big Data Ecosystem'.
Notes
References
| (1) |
M. Assante, L. Candela, D. Castelli, G. Coro, L. Lelii, P. Pagano. Virtual Research Environments as-a-Service by gCube. 8th International Workshop on Science Gateways (IWSG 2016) https://doi.org/10.7287/peerj.preprints.2511v1 |
| (2) |
Bartling, S., & Friesike, S. (2014). Towards another scientific revolution. In Opening Science (pp. 3-15). Springer International Publishing. https://doi.org/10.1007/978-3-319-00026-8_1 |
| (3) |
Michael R. Berthold and Nicolas Cebron and Fabian Dill and Thomas R. Gabriel and Tobias Kotter and Thorsten Meinl and Peter Ohl and Christoph Sieb and Kilian Thiel and Bernd Wiswedel. KNIME: The Konstanz Information Miner. Book Springer, Studies in Classification, Data Analysis, and Knowledge Organization (GfKL 2007). ISBN 978-3-540-78239-1 |
| (4) |
SAS Institute Inc. 2011. Base SAS® 9.3 Procedures Guide. Cary, NC: SAS Institute Inc. Base SAS® 9.3 Procedures Guide. Copyright © 2011, SAS Institute Inc., Cary, NC, USA. ISBN 978-1-60764-895-6. |
| (5) |
Unified Modeling Language User Guide, The (2 ed.). Addison-Wesley. 2005. p. 496. ISBN 0321267974. (See the sample content; look for history.) |
| (6) |
W. M. P. van der Aalst and A. H. M. ter Hofstede. "YAWL: yet another workflow language". Journal Information Systems archive. Volume 30 Issue 4, June 2005. Pages 245-275. https://doi.org/10.1016/j.is.2004.02.002 |
| (7) |
L. Candela, D. Castelli & P. Pagano Virtual Research Environments: An Overview and a Research Agenda. Data Science Journal. 12, pp.GRDI75-GRDI81. https://doi.org/10.2481/dsj.GRDI-013 |
| (8) |
L. Candela, D. Castelli, A. Manzi & P. Pagano. Realising Virtual Research Environments by Hybrid Data Infrastructures: the D4Science Experience. In International Symposium on Grids and Clouds (ISGC) 2014, 23-28 March 2014, Academia Sinica, Taipei, Taiwan, PoS(ISGC2014)022, Proceedings of Science, 2014. |
| (9) |
Stefano Ceri, Themis Palpanas, Emanuele Della Valle, Dino Pedreschi, Johann-Christoph Freytag, Roberto Trasarti: Towards mega-modeling: a walk through data analysis experiences. SIGMOD Record 42(3): 19-27 (2013) https://doi.org/10.1145/2536669.2536673
|
| (10) |
Oinn, T., Addis, M., Ferris, J., Marvin, D., Senger, M., Greenwood, M., ... & Li, P. (2004). Taverna: a tool for the composition and enactment of bioinformatics workflows. Bioinformatics, 20(17), 3045-3054. https://doi.org/10.1093/bioinformatics/bth361 |
| (11) |
LeVeque, R. J., Mitchell, I. M., & Stodden, V. (2012). Reproducible research for scientific computing: Tools and strategies for changing the culture. Computing in Science and Engineering, 14(4), 13. |
| (12) |
Xavier Llora, Bernie XavierLlora, Bernie Acs, Loretta S. Auvil, Boris Capitanu, Michael E. Welge, and David E.Goldberg. 2008. Meandre: Semantic-Driven Data-Intensive Flows in the Clouds. In Proceedings of the 2008 Fourth IEEE International Conference on eScience (e-Science '08). IEEE Computer Society, Los Alamitos, CA, USA, 238-245. https://doi.org/10.1109/eScience.2008.172 |
| (13) |
Ludäscher B., Altintas I., Berkley C., Higgins D., Jaeger-Frank E., Jones M., Lee E., Tao J., Zhao Y. 2006. Scientific Workflow Management and the Kepler System. Special Issue: Workflow in Grid Systems. Concurrency and Computation: Practice & Experience 18(10): 1039-1065. https://doi.org/10.1002/cpe.994 |
| (14) |
JJaval Business Process Management (jBPM). JBoss Community, Red Hat, Inc. Cur. Vers 6.4.0. |
| (15) |
Becker, Jörg, Michael Rosemann, and Christoph Von Uthmann. "Guidelines of business process modeling." Business Process Management. Springer Berlin Heidelberg, 2000. 30-49. https://doi.org/10.1007/3-540-45594-9_3 |
| (16) |
Weerawarana, S., Curbera, F., Leymann, F., Storey, T., & Ferguson, D. F. (2005). Web services platform architecture: SOAP, WSDL, WS-policy, WS-addressing, WS-BPEL, WS-reliable messaging and more. Prentice Hall PTR. |
| (17) |
Andrienko, Natalia, Andrienko, Gennady, Fuchs, Georg, Jankowski, Piotr. Scalable and privacy-respectful interactive discovery of place semantics from human mobility traces. https://doi.org/10.1177/1473871615581216 |
| (18) |
Trasarti, R., Guidotti, R., Monreale, A., Giannotti, F. (2015). MyWay: Location prediction via mobility profiling. Information Systems Journal. https://doi.org/10.1016/j.is.2015.11.002 |
About the Authors
Leonardo Candela is a researcher at the Networked Multimedia Information Systems (NMIS) Laboratory of the Institute of Information Science and Technologies - Italian National Research Council (ISTI — CNR). Dr. Candela graduated with a degree in Computer Science in 2001 at University of Pisa and completed a Ph.D. in Information Engineering in 2006 at the University of Pisa. He joined the NeMIS Laboratory in 2001. Since then he has been involved in various EU-funded projects including CYCLADES, Open Archives Forum, DILIGENT, DRIVER, DELOS, D4Science, D4Science-II, DL.org, EUBrazilOpenBio, iMarine, ENVRI. He was a member of the DELOS Reference Model Technical Committee and of the OAI-ORE Liaison Group. He is currently involved in the D4Science, D4Science-II and DL.org projects. Currently, he is the Project Manager of the BlueBRIDGE Project and CNR lead person in the ENVRIPlus one. His research interests include Data Infrastructures, Virtual Research Environments, Data Publication, Open Science, Digital Library (Management) Systems and Architectures, Digital Libraries Models, Distributed Information Retrieval, and Grid and Cloud Computing.
Fosca Giannotti is a senior researcher at the Information Science and Technology Institute of the National Research Council at Pisa, Italy, where she leads the Knowledge Discovery and Data Mining Laboratory — KDD LAB — a joint research initiative with the University of Pisa, founded in 1995, one of the earliest European research groups specifically targeted at data mining and knowledge discovery. Her current research interests include data mining query languages, knowledge discovery support environment, web-mining, spatio-temporal reasoning, spatio-temporal data mining, and privacy preserving data mining. She has been involved in several research projects both at national and international level, holding both management and research positions. She has been the coordinator of various European and national research projects and she is currently the co-ordinator of the FP6-IST project GeoPKDD: Geographic Privacy-aware Knowledge Discovery and Delivery. She is responsible for the Working Group on Privacy and Security in Data mining of the KDUBIQ network of excellences. She has taught classes on databases and data mining at universities in Italy and abroad. She is the author of more than one hundred publications and served in the scientific committee of various conferences in the area of Logic Programming, Databases, and Data Mining. In 2004 she co-chaired the European conference on Machine Learning and Knowledge Discovery in Data Bases ECML/PKDD 2004. She is the co-editor of the book "Mobility, Data Mining and Privacy", Springer, 2008.
Valerio Grossi holds a Ph.D. in Computer Science from the University of Pisa and he is currently a research fellow at the Department of Computer Science, University of Pisa. He has been a researcher at Department of Pure and Applied Mathematics, University of Padova. His research interests focus on the analysis of massive and complex data including mining data streams, ontology-driven mining, business intelligence and knowledge discovery systems. He took part in several European research projects as a member of the UNIPI research group, among which figure BRITE (Business Register Interoperability throughout Europe), MUSING (MUlti-industry, Semantic-based next generation business INtelliGence) and ICON (Inductive Constraint Programming), where he focused on the development of brand new data mining and knowledge discovery applications and on the research for the development of new business intelligence approaches.
Paolo Manghi is a (PhD) Researcher in computer science at Istituto di Scienza e Tecnologie dell'Informazione (ISTI) of Consiglio Nazionale delle Ricerche (CNR), in Pisa, Italy. He is acting as technical manager and researcher for the EU-H2020 infrastructure projects OpenAIRE2020, SoBigData.eu, PARTHENOS, EOSC, and RDA Europe, and he is the coordinator of the OpenAIRE-Connect project. He is active member of a number of Data Citation and Data Publishing Working groups of the Research Data Alliance; and invited member of the advisory boards of the Research Object initiative. His research areas of interest are today data e-infrastructures for science and scholarly communication infrastructures, with a focus on technologies supporting open science publishing, i.e. computational reproducibility and transparent evaluation of science.
Roberto Trasarti was born in 1979 in Italy. He graduated in Computer Science in 2006, at the University of Pisa. He discussed his thesis on ConQueSt: a Constraint-based Query System aimed at supporting frequent patterns discovery. He started the Ph.D. in Computer Science at the School for Graduate Studies "Galileo Galilei", (University of Pisa). In June 2010 he received his Ph.D. presenting the thesis entitled "Mastering the Spatio-Temporal Knowledge Discovery Process". He is currently a member of ISTI-CNR, and also a member of Knowledge Discovery and Delivery Laboratory. His interests regard Data mining, Spatio-Temporal data analysis, Artificial intelligence, Automatic Reasoning.