D-Lib Magazine
January/February 2017
Volume 23, Number 1/2
Table of Contents
Towards Reproducibility of Microscopy Experiments
Sheeba Samuel
Institute for Computer Science, Friedrich Schiller University Jena
sheeba.samuel@uni-jena.de
Frank Taubert
Institute for Computer Science, Friedrich Schiller University Jena
frank.taubert@uni-jena.de
Daniel Walther
Institute for Computer Science, Friedrich Schiller University Jena
daniel.walther@uni-jena.de
Birgitta König-Ries
Institute for Computer Science, Friedrich Schiller University Jena
Michael Stifel Center Jena for Data-driven and Simulation Science
birgitta.koenig-ries@uni-jena.de
H. Martin Bücker
Institute for Computer Science, Friedrich Schiller University Jena
Michael Stifel Center Jena for Data-driven and Simulation Science
martin.buecker@uni-jena.de
Corresponding Author: Sheeba Samuel, sheeba.samuel@uni-jena.de
https://doi.org/10.1045/january2017-samuel
Abstract
The rapid evolution of various technologies in different scientific disciplines has led to the generation of large volumes of high dimensional data. Studies have shown that most of the published work is not reproducible due to the non-availability of the datasets, code, algorithm, workflow, software, and technologies used for the underlying experiments. The lack of sufficient documentation and the deficit of data sharing among particular research communities have made it extremely difficult to reproduce scientific experiments. In this article, we propose a methodology enhancing the reproducibility of scientific experiments in the domain of microscopy techniques. Though our approach addresses the specific requirements of an interdisciplinary team of scientists from experimental biology to store, manage, and reproduce the workflow of their research experiments, it can also be extended to the requirements of other scientific communities. We present a proof of concept of a central storage system that is based on OMERO (Allan et al., Nature Methods 9, 245-253, 2012). We discuss the criteria and attributes needed for reproducibility of microscopy experiments.
Keywords: Reproducibility, Scientific Experiments, Microscopy Techniques, Data Provenance, Scientific Workflow
1 Introduction
Various emerging methodologies and technologies in different fields of science have led to an increase of experiments conducted in scientific communities. One example for this trend is microscopy. The rapid advancement of technology in this field and the development of new microscopy techniques have tremendously improved the visualization of biological processes and features of the microscopic world at a nanometer resolution. Examples of these powerful techniques include super-resolution microscopy [1], tip-enhanced Raman spectroscopy [2], confocal patch-clamp fluorometry [3] and fluorescence correlation spectroscopy [4]. These new techniques promise new scientific insights, but also bring new challenges with respect to reproducibility of results. First, for many of these techniques, the sheer volume of data is overwhelming and new methods are needed allowing for proper management of this large volume of data. Second, while the microscopes record certain parameters of the experimental settings automatically, there is more information that needs to be recorded to make the experiment reproducible. For instance, it is necessary not only to record experimental protocols, measurements, and images, but also metadata as well as information about the materials used in the experiments. Third, for most methods, the raw data produced by the microscopes need to be processed to obtain the desired final result. For some techniques, this processing involves generating an image out of a series of measurements provided by the microscope. For others, it means combining the image data produced by the microscope with other measurements like electrical current or voltage that are taken simultaneously by additional instruments. For reproducible experiments, these processing steps need to be documented as well.
The new contribution of this paper is the outline of an approach for a software environment that meets these requirements of researchers from experimental biomedicine collaborating within a large-scale project. In this paper, we will introduce the requirements towards data management arising in this setting (Section 2) and their linkage to reproducibility (Section 3). We will then discuss the state of the art (Section 4) and provide an overview of our approach and a brief description of our first prototype (Section 5). We conclude the paper with an outlook on future work (Section 6).
2 Requirement Analysis
The need to design an environment facilitating reproducibility for microscope experiments arises from the Collaborative Research Center ReceptorLight which started in 2015. In this research initiative, scientists from two universities, two university hospitals, and a non-university research institute work together in twenty subprojects to better understand the role and function of membrane receptors by using and advancing high-end light microscopic methods like the ones mentioned above. Collaborative Research Centers (CRCs) are funded by the German Science Foundation for a maximum of three consecutive four-year funding periods.
It is our responsibility to provide a data management platform for the CRC that safekeeps the data produced in the individual subprojects, allows sharing of data among subprojects, and supports data reuse by other CRC scientists in any funding period, as well as by the scientific community beyond the overall funding term of the CRC. A major requirement in this setting is that data are managed in such a way that the scientific experiments associated with that data are reproducible.
The current practice in this community does not necessarily guarantee this. Interviews with our collaborators in the CRC showed that most of the researchers in that scientific community still use the conventional way of writing down the results of the experiments in their (analog) lab notebooks. Very few of them use digital media for storing the information regarding their experiments. The methods used in the CRC require a complex data processing workflow. As an example, let us provide some details on the workflow involved in a confocal patch clamp fluorometry (cPCF) experiment. The patch clamp technique [5] uses a pipette to isolate a single or multiple ion-channels of a cell and measures a current to gain insight in the workings of these channels. The workflow of such an experiment needs the acquisition of images while measuring a current. The images generated through this technique need to have a link to the other data and measurements of the experiment.
Figure 1 shows the deployment of the experimental setup conducted by the biological scientists. The experiment uses different devices and software of the device manufacturer. One of the main instruments is the microscope system which integrates various parts. The system includes microscope stand, illumination sources like lamps and lasers to illuminate the sample, as well as detectors like cameras and photomultipliers. These parts are controlled by software and all settings are stored in the proprietary data format of the microscope manufacturer. In addition, there are several settings of external devices attached to the microscope like external lasers, lifetime measurements, intensity at the interface to the microscope, and intensity in the focal plane. Usually, these additional parts are provided by the biological scientist separately from the microscope. Consequently, they are not integrated into the software of the microscope manufacturer and all settings have to be documented by the scientist with an extra effort.
The electrophysiological device measures voltage or current pulses to clamp the cell membrane potential to a set value or stimulate a cell, capacitance compensation and gain/offset for the measured data. The experiment requires perfusion systems which record the various solutions used in the experiment like bath solution, pipette solution, and transfection solution. The experimental conditions have to be documented as well.
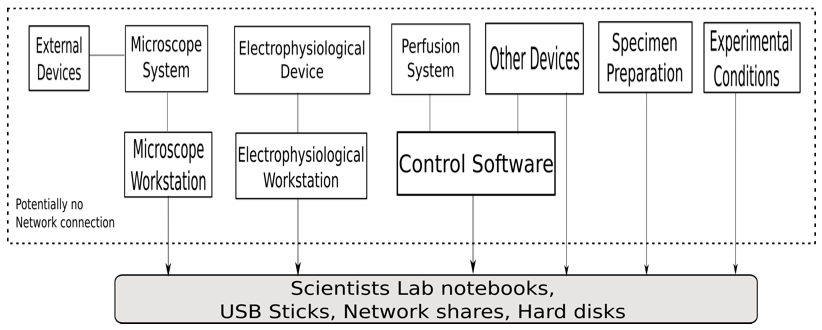
Figure 1: The deployment diagram for the experimental setup by the biological scientists.
Currently, data obtained from different devices are stored on various computers. Also, the data and files received from these computers have to be synchronized in date and time. Most of the workstations associated with these devices do not have an Internet connection for security reasons. Thus, data transfer is done via USB sticks or disks. These storage media are also used for data preservation.
3 Repeatability and Reproducibility of Experiments
The setup described above has a few obvious drawbacks with respect to good data management and reproducibility of experiments. Before we discuss these disadvantages, we need to define the meaning of reproducibility.
According to [6], the term repeatability of a scientific experiment refers to the capability of getting the same (or close-by) specific quantity subject to measurement whenever that experiment is carried out by the same experimenter using the same conditions of measurement. Reproducibility, on the other hand, is more stringent. It refers to the capability of getting the same (or close-by) specific quantity whenever that experiment is carried out by an independent experimenter using different conditions of measurement. Here, the changed conditions of measurement may include the method, location, or time of measurement.
Scientific experiments should be repeatable and reproducible [7].
Several studies suggest that around 10% of published science articles are reproducible [8]. Scientists at the pharmaceutical company Bayer used several approaches to reproduce the published results by collecting data from 67 projects [9] from the field of oncology, cardiovascular system, and women's health. For only 14 of these projects, the published results were reproducible. In the remaining projects, there were inconsistencies between the published results and the in-house findings of the scientists at Bayer.
Studies conducted by the biotech company Amgen reveal that only 6 of the 53 studies in cancer research could be reproduced [10]. These studies identify a serious problem that needs to be tackled by the scientific community. A scientific result can only be considered valid, and of value to the scientific community and society, if it is reproducible.
Goecks et al. [11] identify different aspects of experiments that need to be taken into consideration to achieve reproducibility. In the following, we link those to the setting in CRC ReceptorLight:
- Data and methods used for the experiment:
This includes the input data required for conducting the experiment and the output data generated from the experiment. Methods used for the experiment include the algorithms, pseudo-code, standards followed in performing the experiment, and the different experimental techniques.
- Workflow:
The scientific workflow captures every step performed in the experiment. It is a representation of a series of computational steps done by the experimenter.
- Provenance:
Provenance provides a detailed description of the process followed in the experiment. It includes information like who created the data and when, who modified the data and the time of modification, the list of processes used to create results, whether two products were derived from the same data, etc. [12] It is also required to track information about each step in the experiment. This information can either be manually captured user input data or automatically captured data like the data generated by the instruments, e.g., device settings or parameters.
- User annotations, labeling and tagging:
User annotations give contextual information on the scientific data. Labeling and tagging helps in future identification of the data. These are useful for search and categorization of the datasets.
- Software used, data technologies and version control:
This includes the software used in various devices and the technologies used in the experiment like the scripting language and data management tools. Version control gives information on the version of the software, instruments and materials used in the experiment [13].
- Experiment environment parameters:
These are the environmental conditions of the labroom while the experiment was conducted. It includes external factors like temperature or humidity of the labroom.
- Data sharing, archiving and distribution:
A data sharing policy is required which specifies which and how data and code can be shared with other researchers and research communities [14]. Distribution represents the standards followed in distributing the results, code and the datasets of the experiment [15]. Since the data generated by the experiment are in the scale of terabytes, the data have to be archived in a central space.
- Data storage:
Data storage is another factor in reproducibility. There are many questions arising on data management that need a proper answer before conducting the experiment, including:
- Is there enough data space for all the data produced in the experiment?
- Is there a central storage system available within the working research group to store the produced data?
- Is the data stored in a secured place without any tampering or modification of the raw data?
- Should all the data be stored or only the results of the experiment?
- Is a small amount of raw data stored enough to generate or legitimate the results?
- Machine-readable integration and documentation:
Currently the researchers store their data in a textual format. Storing data and metadata in a machine-readable format makes it easier to trace the experiment. Documentation of all information regarding an experiment makes it possible to rerun the experiment and reanalyze the results.
4 Related Work
In the previous section, we identified the information that needs to be maintained to ensure reproducibility of experiments in the CRC ReceptorLight. In this section, we will discuss existing platforms for data management and how well they meet our requirements. More precisely, we take a look at three related aspects, namely scientific workflow management systems, provenance, and traceability.
A scientific workflow management system [16] is a workflow management system to capture a series of computational steps conducted in a scientific experiment. There are various scientific workflow systems. Some of them are general purpose while others are specific to certain fields like bioinformatics, biodiversity or astronomy. The workflow management system allows scientists to design, create, model, visualize, implement, execute and rerun their analysis and experiment. Some workflow systems like the CoG Kit's Karajan [17] provide a scripting environment to perform more complex actions. Such tools are difficult to use for scientists who have little knowledge in programming languages and computer science technologies. Scientific Workflow Systems like Kepler [18], Vistrails [19] and Triana [20] provide complex composition tools for building workflow in the form of a graph where the nodes represent tasks and edges represent dependencies between tasks. Few of them provide the facility to track, manage, and search provenance data like Vistrails, Triana and Karma [21], while others have the ability to capture and store provenance data. The overview [22] shows that workflow systems are designed for desktop [23], web or cloud.
In [24], Astakhov et al. describe work to adapt the Kepler workflow engine to support "end-to-end" workflows utilizing the data coming from the microscopes installed at the National Center for Microscopy Imaging Research (NCMIR). The system consists of three components: Cell Centered Database, iRODS and Kepler scientific workflow system. The Cell Centered Database manages the data flow. It is a web accessible database for high-resolution 2D, 3D and 4D data obtained from confocal, multiphoton, light and electron microscopy. The system iRODS (i Rule Oriented Data System) is used to store, maintain, archive and provide access to the data obtained from NCMIR microscopes. It allows the distribution of data among available disc spaces in NCMIR. Kepler allows scientists to create workflows to model and perform complex analyses on their scientific data. The prototype is developed mainly for NCMIR projects.
A similar approach by Abramson et al. [25] discusses the extension of Grid workflow engines to perform scientific experiments on virtual microscopes. They have designed and implemented new Kepler actors for accessing microscopes regardless of their physical location, actors that integrate user actions into the workflow and actors that display workflow results on tiled display walls. The work demonstrates the use of scientific workflow systems in specifying and executing the environment. The paper points out the limitation of reusing and sharing the workflows because of the large number of domain specific parameters.
In [26], Pimentel describes the work of combining the noWorkflow [27] and YesWorkflow [28] to capture, store, query and visualize the provenance of results generated by scripts written by researchers. The tool noWorkflow captures provenance information from the program structure and from the key events observed during the execution of scripts. YesWorkflow extracts provenance data from annotation comments made by the authors in the script. The work presents a joint model where the common entities from both of the provenance models were mapped together and provenance information represented by each system can be queried jointly.
In PROV-O [29], the PROV Data Model (PROV-DM) [30] is expressed using the OWL2 Web Ontology Language (OWL2) [31]. It provides a set of classes and properties to represent provenance information in various domains and systems. It can be used to create new classes and properties to model provenance information for different contexts.
The approaches mentioned in this section are based on workflow systems that seem to be too complex for the scientists from the biological domain. Even though some of these workflow systems are rich in features, they are difficult for the scientists to use [32]. Scientists require an infrastructure which is easy to use and where they can easily transition from their conventional way of recording data to the digital form. To get the advantage of provenance and reproducibility of microscopic experiments, the data from the scientists have to be modeled, captured and stored in a central storage system. Sharing of the data should be governed by a set of permissions, as is best practice in many computer science systems. It should also provide the capability to organize the data in such a way that the scientist can relate all the information associated with his or her experiment. From our first preliminary experiences, the prototypical implementation of a new environment presented in the following section is capable of capturing and organizing the experimental data for the scientists working on microscopy techniques.
4 Current Prototype and First Results
We propose an approach where the biological scientists can document all necessary information regarding their experiments while performing them. The researcher can record information of an experiment, the standard operating procedures, molecular biological data, and information about materials used in the experiment. The standard operating procedures include information on the preparation of cells, the transfection/incubation, the synthesis of chemical compounds, etc. The molecular biology data include information about materials like vectors [33], plasmids [34] and proteins [35] used in the experiments.
Figure 2 displays the proposed approach for the microscopy experiments. The data model consists of various input classes representing different types of information regarding an experiment including the classes Experiment, Plasmid, Protein, Chemical Substance and Vector. The Experiment class is the main class that contains all information about a single experiment and includes links to all other classes. While a new Experiment object is created with each experiment, all other classes represent materials usable in various experiments. These classes are referenced in multiple experiments. The approach thus allows the creation of a well-documented set of reusable plasmids, proteins, chemical substances and vectors. This information can then be shared with and improved by other scientists to facilitate the availability of correct information when recording the experimental conditions. The system is also designed to be extensible so that additional classes representing different aspects can be integrated with only modest effort.
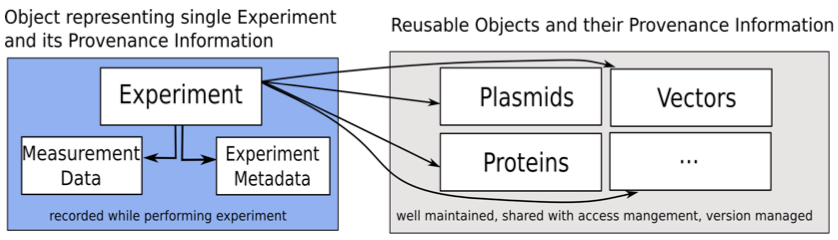
Figure 2: The proposed approach for the microscopy experiments.
Figure 3 shows the setup of our proposed system. The current prototype is developed based on OMERO [36]. The environment OMERO helps the user to store, organize, view, analyze and share data generated by any microscopy experiment. It allows working with the images through a web client, a desktop application (Windows, Mac or Linux) or from 3rd party software. It is being developed by the Open Microscopy Environment consortium, an open source community located mainly in Europe and the USA.
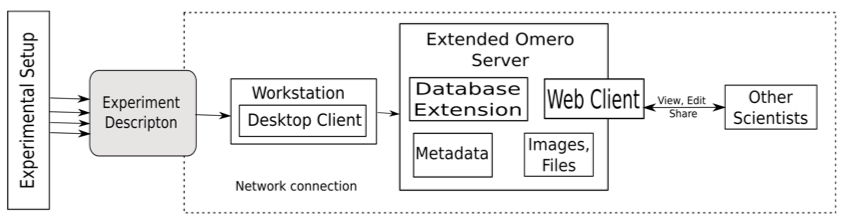
Figure 3: Experimental setup of our proposed system.
In OMERO, data are organized in a hierarchical manner. The hierarchy includes projects, datasets and images. A project consists of several datasets. The datasets can include several images associated with it. OMERO collects the metadata belonging to an image and displays them. Also, the user can add tags, comments and annotations to datasets, images and projects.
In addition to the rich set of features that OMERO already provides, we extended the OMERO server to include the data specified in our approach. This allows the scientist to easily record all the data needed to reproduce his or her experiments. The OMERO server database was extended to include the classes proposed in our approach. We also extended the OMERO web client to include an "Experiment" tab which provides a place to document and view all the information regarding the experiment. Figure 4 shows the OMERO webclient with the extension. A desktop client was developed to deploy the new system on workstations where an Internet connection is not available. So, when a researcher is conducting an experiment, he or she can input all the data in the desktop client. He or she can then upload the images, files and measurements obtained from the devices during the experiment to the server when an Internet connection is available.
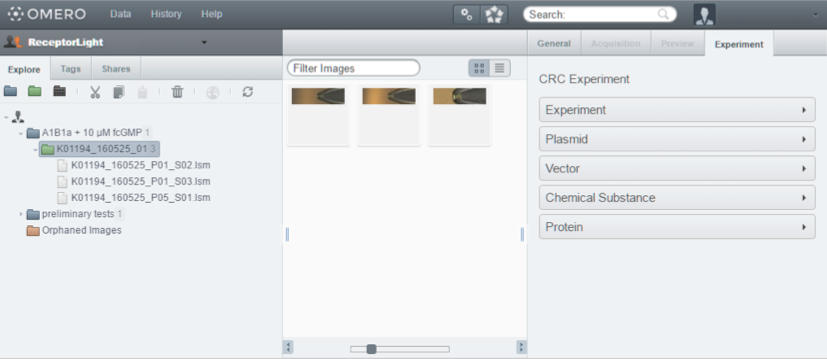
Figure 4: OMERO webclient with the "Experiment" tab extension.
This way the prototype helps the biological scientist to document the experimental protocol, environment and performed steps at the time he or she is performing an experiment, thus minimizing the loss of data naturally occurring when recording these things from memory. All measurements obtained from different devices are stored in the prototype in such a way that all measurements belonging to one experiment can be retrieved and correlated easily. The prototype makes it easier for the scientists to view the images and data of the experimental protocol at one place organized in a hierarchical manner. The prototype also provides access control by providing its users a set of different roles and permissions to restrict modification of data. The groups in the OMERO platform enable sharing of data between users. The version management of the scientific data in the system allows scientists to track modifications by other researchers of his group, correct mistakes and revert back to previous versions if needed.
Currently, the results are manually validated by a group of biological scientists. The system is being continuously extended and improved with the feedback received from the scientists.
6 Future Work and Conclusion
Reproducibility of scientific experiments is very significant for biological scientists to ensure validation and correctness of their work. Each measurement of an experiment may generate gigabytes of data. Thus, there is a high demand for techniques that support the data management related to these experiments. Using a software platform supporting reproducibility, biological scientists can spend more time on their biological work rather than focusing on writing scripts and worrying about the storage of their huge data. The proposed approach provides a central infrastructure for organizing, storing and documenting the data produced by each experiment. This platform is specifically designed keeping in mind the requirements of biological scientists working in the field of microscopy. Data and metadata managed by this platform can be shared among researchers in the same or in different groups. A client-server architecture allows entering data locally on a client without an Internet connection and synchronizing the local copy with the server when an Internet connection is available.
At the time of writing, a set of attributes was added to the platform based on the requirements of a sample group of biological scientists. More attributes will be added to this set gradually, based on the specific requirements of further working groups of biological scientists. Future work will provide the platform to represent the data entered by the scientists in a machine-readable format. It will include representation of data in ontologies and semantic format. The platform will also be integrated with existing tools to export the provenance information from the system. In addition, the prototype will be integrated with a workflow engine to represent the scientific workflow of experiments. More effort is also needed to document the functionality of the platform. With a sufficiently detailed documentation, the current platform will enable a biological scientist to automate the data analysis in such a way that he or she selects only the data sets and the analysis procedure to be included in that analysis.
Acknowledgements
This research is partially supported by the "Deutsche Forschungsgemeinschaft" (DFG) in Project Z2 of the CRC/TRR 166 "High-end light microscopy elucidates membrane receptor function—ReceptorLight". We thank Christoph Biskup and Kathrin Groeneveld from the Biomolecular Photonics Group at University Hospital Jena, Germany, for providing the requirements to develop the proposed approach and validating the system.
References
| [1] |
B. Huang, M. Bates, and X. Zhuang. "Super resolution fluorescence microscopy." Annual Review of Biochemistry 78 (2009): 993-1016. https://doi.org/10.1146/annurev.biochem.77.061906.092014 |
| [2] |
V. Deckert. "Tip-Enhanced Raman Spectroscopy." Journal of Raman Spectroscopy 40 (2009): 1336-1337. https://doi.org/10.1002/jrs.2452 |
| [3] |
C. Biskup et al. "Relating ligand binding to activation gating in CNGA2 channels." Nature 446 (2007): 440-443. https://doi.org/10.1038/nature05596 |
| [4] |
R. Rigler and E. S. Elson, eds. Fluorescence correlation spectroscopy: Theory and Applications. Vol. 65. Springer Science & Business Media, 2012. |
| [5] |
O. P. Hamill, A. Marty, E. Neher, B. Sakmann, and F. Sigworth. "Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches." Pflügers Archiv 391 (1981): 85-100. |
| [6] |
B. N. Taylor and C. E. Kuyatt, eds. Guidelines for Evaluating and Expressing the Uncertainty of NIST Measurement Results, (NIST Tech. Note 1297, U.S. Government Printing Office, Washington, D.C.), 1994. |
| [7] |
A. Casadevall and F.C. Fang. "Reproducible science." Infection and Immunity 78 (2010): 4972-4975. https://doi.org/10.1128/IAI.00908-10 |
| [8] |
M. Baker. "1,500 scientists lift the lid on reproducibility." Nature 533 (2016): 452-454. https://doi.org/10.1038/533452a |
| [9] |
F. Prinz, T. Schlange, and K. Asadullah. "Believe it or not: How much can we rely on published data on potential drug targets?" Nature Reviews Drug Discovery 10 (2011): 712-713. https://doi.org/10.1038/nrd3439-c1 |
| [10] |
C. G. Begley and L. M. Ellis. "Drug development: Raise standards for preclinical cancer research." Nature 483 (2012): 531-533. https://doi.org/10.1038/483531a |
| [11] |
J. Goecks, A. Nekrutenko, and J. Taylor. "Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences." Genome Biology 11 (2010): 1-13. https://doi.org/10.1186/gb-2010-11-8-r86 |
| [12] |
M. Greenwood, C. Goble, R. Stevens, J. Zhao, M. Addis, D. Marvin, L. Moreau, and T. Oinn, "Provenance of e-Science Experimentsexperience from Bioinformatics." In: Proceedings of the UK OST e-Science 2nd AHM, 2003. |
| [13] |
R. Koenker and A. Zeileis. "On Reproducible Econometric Research." Journal of Applied Econometrics, 24 (2009): 833-847. https://doi.org/10.1002/jae.1083 |
| [14] |
R. D. Peng. "Reproducible Research in Computational Science." Science 334 (2011): 1226-1227. https://doi.org/10.1126/science.1213847 |
| [15] |
R. D. Peng, F. Dominici, and S. L. Zeger. "Reproducible Epidemiologic Research." American Journal of Epidemiology 163 (2006): 783-789. https://doi.org/10.1093/aje/kwj093 |
| [16] |
V. Curcin and M. Ghanem. "Scientific workflow systems—can one size fit all?" In: Proceedings of 2008 Cairo International Biomedical Engineering Conference (CIBEC). IEEE (2008). https://doi.org/10.1109/CIBEC.2008.4786077 |
| [17] |
G. von Laszewski and M. Hategan, "Java CoG Kit Karajan/GridAnt Workflow Guide", Technical Report, Argonne National Laboratory, Argonne, IL, USA, 2005. |
| [18] |
B. Ludäscher, I. Altintas, C. Berkley, D. Higgins, E. Jaeger, M. Jones, E. A. Lee, J. Tao, and Y. Zhao. "Scientific workflow management and the Kepler system." Concurrency and Computation: Practice and Experience 18 (2006): 1039–1065. https://doi.org/10.1002/cpe.994 |
| [19] |
S. Callahan, J. Freire, E. Santos, C. Scheidegger, C. Silva, and H. Vo. "VisTrails: Visualization meets data management." In: Proceedings of the 2006 ACM SIGMOD International Conference on Management of Data, 2006, pp. 745–747. |
| [20] |
I. Taylor, M. Shields, I. Wang, and A. Harrison, "Visual grid workflow in Triana." Journal of Grid Computing 3 (2005): 153-169. |
| [21] |
Y. L. Simmhan, B. Plale, and D. Gannon. "Performance evaluation of the Karma provenance framework for scientific workflows." In: International Provenance and Annotation Workshop, IPAW, Springer, Berlin, 2006. |
| [22] |
E. Deelman, D. Gannon, M. Shields, and I. Taylor. "Workflows and e-Science: An overview of workflow system features and capabilities." Future Generation Computer Systems 25 (2009): 528-540. |
| [23] |
T. Oinn, M. Addis, J. Ferris, D. Marvin, M. Senger, M. Greenwood, T. Carver, K. Glover, M.R. Pocock, A. Wipat and P. Li. "Taverna: A Tool for the Composition and Enactment of Bioinformatics Workflows." Bioinformatics 20 (2004): 3045-3054. https://doi.org/10.1093/bioinformatics/bth361 |
| [24] |
V. Astakhov et al. "Prototype of Kepler Processing Workflows for Microscopy and Neuroinformatics." Procedia Computer Science 9 (2012): 1595-1603. |
| [25] |
D. Abramson et al. "Virtual microscopy and analysis using scientific workflows." In: Proceedings Fifth IEEE International Conference on e-Science, 2009. |
| [26] |
J. F. Pimentel et al. "Yin & Yang: Demonstrating complementary provenance from noWorkflow & YesWorkflow." In: M. Mattoso and B. Glavic (eds.) Provenance and Annotation of Data and Processes. LNCS, vol. 9672, pp. 161-165. Springer (2016). https://doi.org/10.1007/978-3-319-40593-3_13 |
| [27] |
L. Murta, V. Braganholo, F. Chirigati, D. Koop, J. Freire. "noWorkflow: Capturing and analyzing provenance of scripts." In: B. Ludäscher and B. Plale (eds.) Provenance and Annotation of Data and Processes. LNCS, vol. 8628, pp. 71-83. Springer (2015). https://doi.org/10.1007/978-3-319-16462-5_6 |
| [28] |
T. McPhillips et al. "YesWorkflow: A user-oriented, language-independent tool for recovering workflow information from scripts." arXiv preprint arXiv:1502.02403 (2015). |
| [29] |
T. Lebo, S. Sahoo, and D. MacGuiness, eds. "PROV-O: The PROV Ontology." W3C Recommendation, version 30. April 2013. World Wide Web Consortium (2013). |
| [30] |
L. Moreau and P. Missier, eds. "Prov-dm: The prov data model." W3C Recommendation, version 30. April 2013. World Wide Web Consortium (2013). |
| [31] |
W3C OWL Working Group, eds. "OWL 2 Web Ontology Language Document Overview." W3C Recommendation, version 11. December 2012. World Wide Web Consortium (2012). |
| [32] |
D. Garijo et al. "Common motifs in scientific workflows: An empirical analysis." Future Generation Computer Systems 36 (2014): 338-351. |
| [33] |
B. Alberts, A. Johnson, J. Lewis et al. Molecular Biology of the Cell. 4th edition. New York: Garland Science; 2002. Isolating, Cloning, and Sequencing DNA. |
| [34] |
H. Lodish, A. Berk, S. L. Zipursky et al. Molecular Cell Biology. 4th edition. New York: W. H. Freeman; 2000. Section 7.1, DNA Cloning with Plasmid Vectors. |
| [35] |
B. Alberts, A. Johnson, J. Lewis et al. Molecular Biology of the Cell. 4th edition. New York: Garland Science; 2002. The Shape and Structure of Proteins. |
| [36] |
C. Allan, J.-M. Burel, J. Moore, C. Blackburn, M. Linkert, S. Loynton, D. MacDonald, W.J. Moore, C. Neves, A. Patterson et al. "Omero: Flexible, model-driven data management for experimental biology." Nature Methods 9 (2012): 245-253. |
About the Authors
Sheeba Samuel is doing PhD in Integrative Data Management and Processing at the University of Jena, Germany. She received her Master of Technology in Information Technology in 2013 at IIIT Bangalore, India. Her current research interests include scientific research data management, reproducibility and semantic web. She is currently working in the DFG project ReceptorLight.
Frank Taubert received his Diploma from the "Carl Zeiss" University in Jena, as well as a MBA from the "Ernst Abbe" University of Applied Science, also in Jena. After working for Zeiss Microscopy as a Software Engineer, he is currently a PhD student at the chair for Advanced Computing at the University of Jena. His current research is on storage of scientific data, reproducibility and big data.
Daniel Walther is a PhD student at the faculty for mathematics and computer sciences at the University of Jena since 2015. He received his Dipl.-Ing. in automation engineering in 2009 and M.Eng. in system design in 2011 at the Ernst Abbe university of applied science in Jena. He used to work for Carl Zeiss Microscopy GmbH from 2011 to 2015 as a software engineer. Currently he is working at methods to improve reproducibility and storing data of microscopy experiments.
Birgitta König-Ries holds the Heinz-Nixdorf Chair for Distributed Information Systems at the University of Jena, Germany. Ever since her PhD, the focus of her work has been on information management and integration in distributed environments. Over the last few years, her group has addressed these problems in the context of research data management in particular in biodiversity and related disciplines. In the frame of several collaborative DFG projects (including the German Centre for Integrative Biodiversity Research (iDiv), Halle-Jena-Leipzig, CRCs AquaDiva and ReceptorLight, the Biodiversity Exploratories and the Jena Experiment), the group coordinates the development of the open source data management platform BExIS 2 and investigates different aspects of using semantic techniques to enhance research data management and integration.
H. Martin Bücker is currently a Full Professor in the Department of Mathematics and Computer Science, Friedrich Schiller University Jena, Jena, Germany, which he joined in 2012. Prior to this, he was a researcher at Forschungszentrum Jülich, a visiting scientist at Argonne National Laboratory, and a senior researcher at RWTH Aachen University. His research focuses on parallel algorithms, high-performance computing, combinatorial scientific computing, and automatic differentiation.